Clara Albiñana
@clara_albi
Followers
380
Following
1K
Media
18
Statuses
158
Postdoctoral researcher @AarhusUni | Currently visiting @IMBatUQ
Brisbane, Queensland
Joined October 2012
Our paired GWAS of vitamin D and its binding protein (DBP) is now published in @NatureComms 🌞🧴 This is the largest GWAS on DBP to date with >65k blood samples from neonates Keep reading this🧵for a summary of our exciting results! https://t.co/BwIvDSx06q
3
27
116
📢REGISTRATION IS NOW OPEN: 4-6th July in York. Join us for the XII International Symposium SRUK/CERU: "Facing Challenges" We will discuss the most pressing research topics, work-life balance, women in STEM and more. Join us! Register now: https://t.co/PeO5Rj6Wmc
3
7
8
Workshop: AI, Data Science, and Genomics Applications Trondheim, Norway – September 9-10, 2024 Will provide participants with conceptual introductions, higher-level framing, and hands-on practicums. Space is limited to 40 participants, so register now! https://t.co/Hh1aV4sZS9
0
6
9
Two-day launch meeting for our @wellcometrust Mental Health award, AMBER, Antidepressants Medications: Biology, Exposure and Response, bring together diverse team across four unis. 1/n
1
7
48
The ways we think about genetic variation, and the terminology we use, should reflect the pertinent scale of worldwide population genomic variation and not just the cohorts that happen to have been sequenced to date. 9/n
3
8
39
10 women in STEM - pick it up and keep it moving! @IsabellBrikell @Xiaoqin_Liu_AU @trinemunkolsen @ZYilmazPhD @LiselotteVP @jessmndy @KMusliner @DemontisDitte @WrayNaomi @Britt_Mitchell1
10 women in STEM - pick it up and keep it moving! @eve_salminen @ttukiainen @HannaMOllila @GenEpi_Sara @AnniqueC
@bnwolford @HujoelM @AmyIsMyNickname @sinarueeger @clara_albi
0
0
7
The correlates of neonatal complement component 3 and 4 protein concentrations with a focus on psychiatric and autoimmune disorders @CellGenomics Funded by @GrundforskFond Niels Bohr Professorship https://t.co/xVKmNsMdE9
1
9
27
Read more about this new *LDpred2* work, now published in @AJHGNews 🥳 https://t.co/3aJKP0jJO7
2
34
122
Is the genetic of disease susceptibility and progression shared? We look at this question within a global biobank collaboration across 10 major diseases 💁
5
44
125
Hi everyone, Our paper about boosting the power of GWAS using polygenic scores (PGS) is just out in @NatureGenet. Please check it out here:
4
50
163
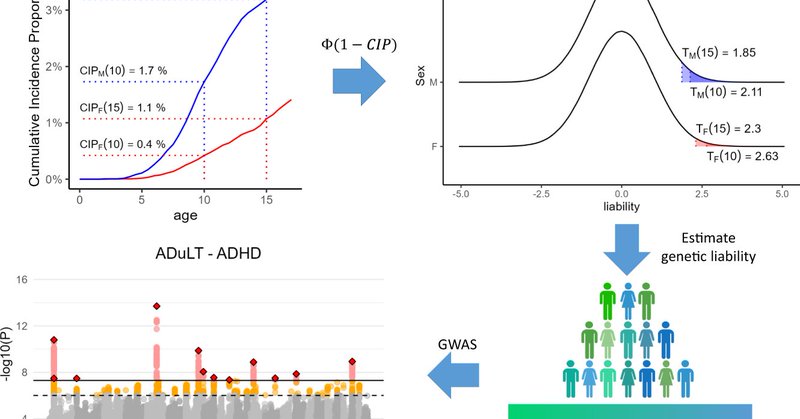
The second paper of my PhD is finally out! Have you ever wanted to account for time-to-event in a GWAS, but not known if you would actually increase power? Then find out here!
nature.com
Nature Communications - Robust genome-wide association study (GWAS) methods that can utilise time-to-event information such as age-of-onset will help increase power in analyses for common health...
0
30
87
When your friends are great scientists, you make a seminar! Join us at Copenhagen University for a top-notch psychiatric genetics speaker line up. Friday, September 15th. 9:00 to 15:30.
2
10
30
📢Great news! We (@iamslambert @minouye271 ) just released pgsc_calc ( https://t.co/WsaPQJjjpW) compatible model files on our OmicsPred portal ( https://t.co/KTTrt7rGkh)! Hopefully, it can make our life easier when calculating thousands of multi-omic genetic scores on a cohort.
1
4
25
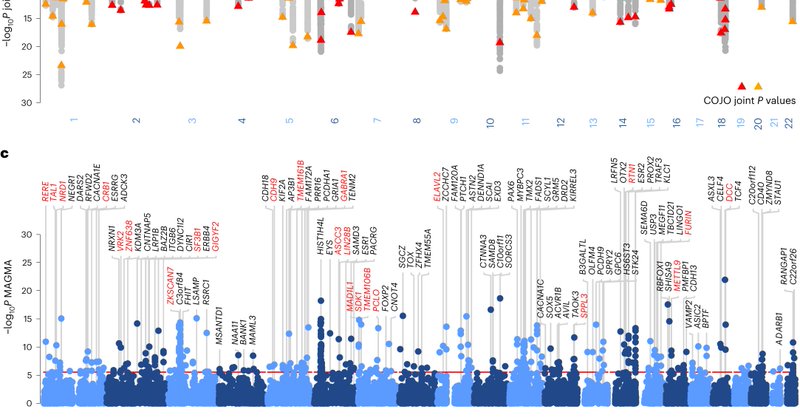
A genome-wide meta-analysis of data from 6 US and European cohorts involving 1.3 million individuals identifies 243 genetic variants associated with risk and pathophysiology of depression @AndersBorglum
https://t.co/5mVa9DlGeW
nature.com
Nature Medicine - A genome-wide meta-analysis of data from six US and European cohorts involving 1.3 million individuals identifies 243 genetic variants associated with risk and pathophysiology of...
1
20
68
If you are here and wanna hear more about our Multi-PGS work come to the G4 Genetics of complex disease symposium on Thursday https://t.co/Y9O2Clnsij
medrxiv.org
The predictive performance of polygenic scores (PGS) is largely dependent on the number of samples available to train the PGS. Increasing the sample size for a specific phenotype is expensive and...
0
0
2
Why do family history and polygenic scores explain independent variance for complex traits? Is it because family history is an 'environmental instrument'? Or are these just two very noisy genetic predictors? See our new paper for a deep dive. https://t.co/oJ6sJJLARk
medrxiv.org
Genetics as a science has roots in studying phenotypes of relatives, but molecular approaches facilitate direct measurements of genomic variation within individuals. Agricultural and human biomedical...
3
33
121
We are excited to share a pre-print of the flagship project of @intervene_eu! https://t.co/Yc73gwchaN
medrxiv.org
Polygenic Scores (PGSs) offer the ability to predict genetic risk for complex disease across the life course; a key benefit over short-term prediction models. To produce risk estimates relevant for...
1
31
68
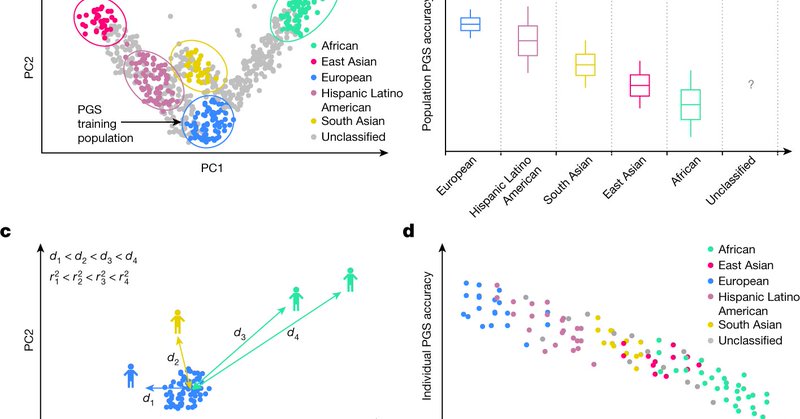
We offer an individual-level interpretation of polygenic score (PGS) transferability across genetic ancestries: PGS accuracy varies individual-to-individual across the genetic-ancestry continuum, even in populations usually labeled as homogeneous. 1/ https://t.co/xqwyKskqbq
nature.com
Nature - Using two large biobank datasets, a study shows that the accuracy of polygenic scores decreases as a function of relatedness at the individual level when modelling genetic ancestry as a...
4
122
299