EmilP
@EmilP94637497
Followers
64
Following
46
Media
1
Statuses
12
The second paper of my PhD is finally out! Have you ever wanted to account for time-to-event in a GWAS, but not known if you would actually increase power? Then find out here!
nature.com
Nature Communications - Robust genome-wide association study (GWAS) methods that can utilise time-to-event information such as age-of-onset will help increase power in analyses for common health...
0
30
87
Dozens of cool PGS methods have been proposed, but which is the best? Fortunately @MennoWitteveen came up with an ingenious approach to answer this using a Public Privacy-preserving Benchmark. Check the poster (# 3566) at 3pm today at #ASHG. Also preprint https://t.co/pYwl1eRECh
2
10
46
Fantastic paper by the clever @clara_albi and the productive team led by @bvilhjal at the National Centre for Register-based Research @AarhusUni
@GrundforskFond @lundbeckfonden Multi-PGS enhances polygenic prediction: weighting 937 polygenic scores
medrxiv.org
The predictive performance of polygenic scores (PGS) in clinical risk models is largely dependent on the number of samples available to train the score, together with the proportion of causal...
1
8
28
Jette Steinbach smashed her talk on family liabilities; supervised by @bvilhjal at @AarhusUni_int supported Emil Pedersen #WCPG2022
0
3
13
It seems @bvilhjal beat me to it. Here is an overview of my new preprint!
It turns out that accounting for age-of-onset information using (efficient) Cox-based GWAS methods may not always result in increased power to detect genetic variants in practice.
0
0
5
Take a look at our new paper @AJHGNews "*Leveraging both individual-level genetic data and GWAS summary statistics increases polygenic prediction*" Squeeze 🍊 the available data to maximize prediction of polygenic risk scores (PRS) Summary 🧵
New! @clara_albi @bvilhjal @privefl & colleagues explore strategies to boost PRS prediction accuracy https://t.co/5M8H0Focd1
1
12
38
In short, we have developed a method that can fine-tune the estimate for a genetic liability per individual. The method can be thought of as a survival model, combining principles of survival analysis models with family history, in a GWAS setting. 4/4
1
0
6
We have implemented LT-FH++ in an R package: https://t.co/qu71tlCqTQ The implementation utilizes a Gibbs sampler and is highly scalable. If you are already familiar with the exellent paper on LT-FH by Margaux Hujoel, we also implemented a *very* fast version of LT-FH! 3/4
github.com
Implementation of LTFH++. Contribute to EmilMiP/LTFHPlus development by creating an account on GitHub.
1
0
4
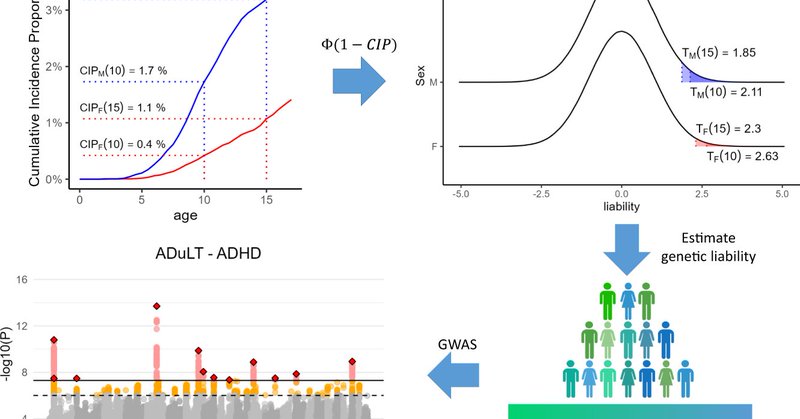
We utilize an age-dependent liability threshold model to get personalized thresholds for every individual, even for the family members. In short, we can account for, e.g. sex, birth year, family history, and age-of-onset on a phenotype level and per individual. 2/4
1
3
5
My first preprint is now on biorxiv! The paper is called: "Accounting for age-of-onset and family history improves power in genome-wide association studies" and the method developed is called "LT-FH++" https://t.co/9U2NQRjHf1 1/4
1
13
43