Mariusz Czarnocki-Cieciura
@MariuszCzC
Followers
261
Following
2K
Media
16
Statuses
588
structural biologist in the Nowotny lab at IIMCB Warsaw
Warszawa, Polska
Joined February 2018
RT @MassSpecVaniLi: 🤩 JOB ALERT 🤩 Come work with me and two cutting-edge LC-MS/MS systems at our brand new Mass Spectrometry Facility @IIMC….
skk.erecruiter.pl
Job offer Mass Spectrometry Technology Specialist, Międzynarodowy Instytut Biologii Molekularnej i Komórkowej w Warszawie, Warszawa
0
10
0
RT @JoePetersLab: Tn7 family transposons that include all guide RNA-directed transposons (CAST) are common in bacteria. We now find diverse….
biorxiv.org
Tn7 family elements are DNA transposons in bacteria that show tight control of targeting, including multiple varieties that evolved guide RNA-directed transposition. We find diverse Tn7 elements...
0
9
0
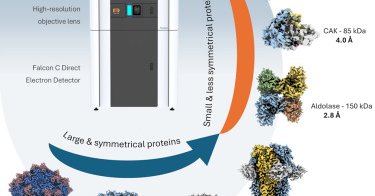
RT @Structure_CP: Online now! Sub-3 Å resolution protein structure determination by single-particle cryo-EM at 100 keV .
cell.com
Karia et al. demonstrate that a 100 keV cryo-TEM system equipped with an XFEG, high-resolution optics, and the Falcon C direct electron detector enables near-atomic resolution structure determination...
0
26
0
RT @biorxiv_biophys: VitriFlex: An Open-Source, Modular, and Customizable Robotic Platform for Cryo-EM Grid Preparation .
biorxiv.org
Specimen preparation remains the principal bottleneck in cryo electron microscopy (cryo EM). Achieving uniformly thin ice without denaturation, preferred particle orientation, or air-water interface...
0
9
0
RT @GlattSebastian: Dear all,. We are starting an exciting project with @ClmentCarr5 in Paris, France. A fully funded PhD position is ava….
0
21
0
RT @A_Chacinska: In this subproject of AMBER we seek to understand the structural basis of mitochondrial protein import. Structural biologi….
0
2
0
RT @mitchguttman: Many proteins bind RNA, yet we still don’t know what RNAs most bind because methods map one RBP at a time. In @CellCellPr….
0
114
0
RT @miroastore: That's a wrap! The results of the first #cryoEM heterogeneity challenge are up on biorxiv!. https://….
0
30
0
RT @iaincheeseman: New preprint! We solve a mystery you didn't know existed. Mitotic cells lack new transcription but require ongoing trans….
biorxiv.org
In the presence of cell division errors, mammalian cells can pause in mitosis for tens of hours with little to no transcription, while still requiring continued translation for viability. These...
0
37
0
RT @LucasFarnung: I am pleased to announce Reactor—the complex formation calculator for multi-component protein complex formation experimen….
0
4
0
RT @EMaleckaPhD: Ready to explore the world of phages through transcriptomics and advanced microscopy? Apply now to join our research team!….
0
7
0
RT @AlexanderSobol6: Happy to share Structural basis of the inhibition of TRPV1 by analgesic sesquiterpenes. In collaboration with León D.….
pnas.org
The Transient Receptor Potential Vanilloid 1 (TRPV1) ion channel is expressed in primary nociceptive afferents, which participate in processes such...
0
9
0
RT @biorxiv_biophys: High-resolution cryo-EM structures of small protein-ligand complexes near the theoretical size limit .
biorxiv.org
Cryo-electron microscopy (cryo-EM) is widely used to determine macromolecular structures at atomic resolution. The theoretical size lower limit of particles for cryo-EM analysis is 38 kDa, limited by...
0
14
0
RT @JZmorzynska: 🚨 Postdoc Opportunity Alert!. I'm currently looking for a postdoc to join our research team! If you're passionate about de….
0
1
0
RT @ZhongingAlong: CryoDRGN-AI ❄️🐉🤖 is now published in @naturemethods!!! So excited to see this out and a huge congrats to @axlevy0 and te….
0
32
0
RT @A_Chacinska: Structural biologists- join us to work on the mechanisms of protein transport to mitochondria! .We work in the interdyscip….
0
6
0
Please check out the newest preprint from our lab (with my small contribution) :).
I am very excited to share our recent preprint in which we are structurally investigating macromolecular complexes that contain DNA-binding proteins ICP8 of Herpes Simplex Virus 1 and BALF2 of the Epstein-Barr Virus.
0
1
6
RT @t_goral: Here it comes! A long-awaited paper led by @MatyldaIzert and @m_gorna is now out. It features multi-mode approach combining di….
0
1
0
RT @GaurVineetSBL: We are looking for a Project Associate-I to work on an exciting DBT-funded project on DNA repair & recombination. If yo….
0
7
0
RT @biorxiv_molbio: Structural insights into DNA annealing and recombination by herpesviral DNA-binding proteins ICP8 and BALF2 https://t.c….
biorxiv.org
The DNA-binding proteins of herpesviruses are key factors involved in multiple stages of DNA replication and recombination. The well-studied ICP8 protein of Herpes Simplex Virus 1 binds single-stra...
0
2
0