The Ding Lab
@DingLab_WashU
Followers
106
Following
21
Media
0
Statuses
26
Lab led by Dr. Li Ding @washumedicine | Pioneering cancer proteogenomics | Affiliated with @WashUOncology and @GenomeInstitute
St. Louis, MO
Joined August 2023
🧬 We are pleased to announce that, as part of our work with #HTAN, three new studies from our lab are published across the @Nature family! These studies offer insights into tumor biology, from spatial dynamics to chromatin accessibility in breast cancer.
nature.com
A collection of research articles, methods and datasets from a collaborative initative tracking human tumor evolution in space and time
1
0
2
Congratulations to Chia-Kuei (Simon) Mo, @ClaraLiu97, @IglesiaMichael, @reykajayasinghe, and many others! Special thanks to our collaborators, including @benjraphael and the dedicated investigators at @WashUOncology, for their invaluable contributions to this work! #HTAN
0
0
0
3️⃣ Inferring Tumor Phylogeography from Spatially Resolved Transcriptomics (@naturemethods): Introducing CalicoST, a method to map copy number aberrations across tumor regions, enabling high-resolution reconstructions of tumor evolution.
nature.com
Nature Methods - This work presents CalicoST for inferring allele-specific copy numbers and reconstructing spatial tumor evolution by using spatial transcriptomics data.
1
0
1
2️⃣ Breast Cancer Subtypes and Lineages Defined by Chromatin Accessibility (@NatureCancer): This study maps chromatin accessibility to reveal lineage-specific dynamics in #BreastCancer, advancing understanding of subtype pathways and therapeutic targets.
nature.com
Nature Cancer - Ding and colleagues use bulk, single-cell and single-nucleus multi-omics together with spatial transcriptomics and multiplex imaging of clinical tumor samples to characterize gene...
1
0
0
1️⃣ Tumor Evolution in 2D and 3D Microenvironments (@Nature): This study uncovers how tumor cells interact with surrounding stromal and immune cells to form distinct “tumor microregions”.
nature.com
Nature - Visium spatial transcriptomics, single-nucleus RNA sequencing and co-detection by indexing are used to identify distinct spatial microregions in tumours and their microenvironment across...
1
0
0
Researchers at WashU Medicine have created 3D maps of tumor structures, revealing how cancer cells and their environment are organized at a single-cell level. The study, published in Nature, could pave the way for innovative therapies. https://t.co/eyfht8czK0
0
3
11
Tumour atlases enable researchers to navigate through cancers
nature.com
Nature - Human tumour atlases have been generated for 21 tissue sites from almost 2,000 individuals, providing insights into cancer evolution.
2
25
82
PRESS RELEASE: New insights from 10 Human Tumor Atlas Network studies provide critical information on how cancer tumors develop, spread, and respond to treatments. The research is to be published tomorrow in @NatureCancer. Learn more: https://t.co/dJWoONoI0M
#CancerMoonshot #HTAN
3
25
55
A new study co-led by #WashUMed researchers analyzes the epigenomes of tumor cells across 11 cancer types, shedding light on the role of epigenetics in cancer formation, growth and spreading. Learn more:
medicine.washu.edu
Epigenetics of cancer critical in understanding tumor initiation, growth and spreading
1
3
7
Thank you to our many collaborators including Feng Chen, PhD @WUSTLmed; Ryan Fields, MD @WashUSurgery; and Ben Raphael , PhD @PrincetonCS—and to their dedicated teams. Additionally, we greatly appreciate the contributing members from @SitemanCenter, @BSC_CNS, and @UCSF_ObGynRS!
0
0
1
A special congratulations to co-first authors Nadya Terekhanova, PhD; Alla Karpova; and Wen-Wei Liang, PhD! @WUSTLmed
1
0
2
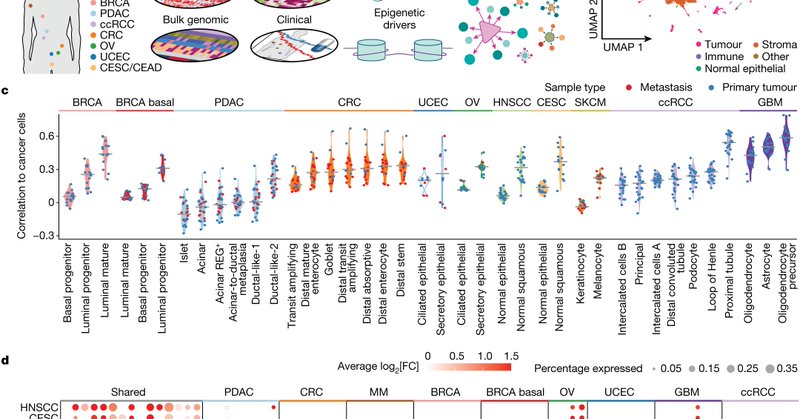
Excited to share that our most recent pan-cancer study is out in @Nature! We uncovered cis- and trans- epigenetic events linked to cancer initiation and progression across 11 cancer types. #PanCancer #HTAN #Epigenetics
https://t.co/IUr8qKzN9b
nature.com
Nature - A pan-cancer epigenetic and transcriptomic atlas identifies epigenetic drivers associated with cancer transitions.
7
28
109
Researchers at #WashUMed, @broadinstitute of @MIT and @Harvard, @BYU and more have completed an analysis of the proteins driving cancer across multiple tumor types. This raises the prospect of new therapies that block key proteins that drive cancer growth.
medicine.washu.edu
Understanding of molecular basis of cancer may lead to new therapies
1
16
61
Today Cell, Cancer Cell and Cell Reports Medicine are pleased to publish the 1st wave of Pan-#Cancer multiomics papers looking at over 1000 patients across 10 #tumor types from @theNCI Clinical Proteomic Tumor Analysis Consortium #CPTAC (1/n) @Cancer_Cell @CellRepMed #openaccess
5
288
767
Data/Resources: Yongchao Dou, PhD, Bing Zhang,PhD @bcmhouston; Felipe Da Veiga Leprevost, PhD, Alexey Nesvizhskii, PhD @umichmedicine;Yifat Geffen, PhD Gad Getz, PhD @broadinstitute ; Anna Calinawan, Pei Wang, PhD @IcahnMountSinai ; Ana Robles, PhD @theNCI; Samuel Payne, PhD @BYU
0
0
0
PTM: Yifat Geffen, PhD, Shankara Anand, Yo Akiyama, Gad Getz, PhD @getz_lab @broadinstitute ; Tomer Yaron, PhD, Lewis Cantley, PhD @CantleyLab @harvardmed @DanaFarber
1
0
0
Driver: Eduard Porta-Pardo, PhD @BSC_CNS; Collin Tokheim, PhD @DanaFarber , Tomer Yaron, PhD, Lewis Cantley, PhD @CantleyLab @harvardmed
@DanaFarber; Chandan Kumar-Sinha, PhD @umichmedicine ; Alexander Lazar, MD, PhD @MDAndersonNews ; Gad Getz, PhD @getz_lab @broadinstitute
1
0
0
#NCI #CPTAC is truly a multi-institutional effort. A special congratulations to all of our collaborators! Deep appreciation to Ana I. Robles, PhD and Henry Rodriguez, PhD, MBA @theNCI for their outstanding leadership in this effort! #TeamScience
1
1
1
Fernanda Martins Rodrigues; Nadezhda V. Terekhanova, PhD; Yige Wu, PhD; and Lijun Yao, PhD. Also extending acknowledgment to our @WUSTLmed collaborator, Milan Chheda, MD! #TeamScience
0
0
0