Ben Raphael
@benjraphael
Followers
2K
Following
174
Media
51
Statuses
287
Professor of Computer Science, Princeton University
Princeton, NJ
Joined March 2014
A great thread from @uthsavc describing some of the math behind GASTON, our new method for modeling spatial variation in gene expression, now out in @naturemethods!
GASTON, our method to learn “topographic maps” of gene expression, is out now @naturemethods! IMO the coolest part is a new model of *spatial gradients in sparse data*. As is typical for bio papers, it’s buried in Methods, but see below for a quick outline on the math 👇
0
1
19
Congratulations to all the winners and finalists of the 2024 @Regeneron Science Talent Search! It was inspiring to meet the next generation of leaders in science and technology!
🏅We're excited to announce the winners of the 2024 @Regeneron Science Talent Search. Congratulations to these extraordinary young scientists! #RegeneronSTS
1
1
13
Thank you @rohitsingh8080 for hosting me at @DukeCellBiology! A great visit with a packed auditorium and many stimulating conversations! This photo was me being delightfully surprised that I was speaking in a lecture series next to these truly distinguished cell biologists
I forgot to take a photo during his talk, so this will have to do. @benjraphael gave a fantastic talk at @DukeCellBiology on the creative and elegant spatial transcriptomics methods from the Raphael Lab. Check out Gaston, Belayer, PASTE2 and the rest!
0
0
13
Also, a quick observation: our 1-D coordinate is a spatial *potential function* - like the height in the famous Waddington's landscape - for some tissues Modeling spatial information -> more accurate differentiation potential vs pseudotime methods which do not use spatial info
0
2
5
GASTON software available at https://t.co/ydHTNhTyan. Tutorials are here with more coming soon https://t.co/Ox6tNbIw7Q (7/7)
github.com
GASTON: deep learning algorithm learns "topography" of a tissue slice from spatial transcriptomics data - raphael-group/GASTON
0
1
5
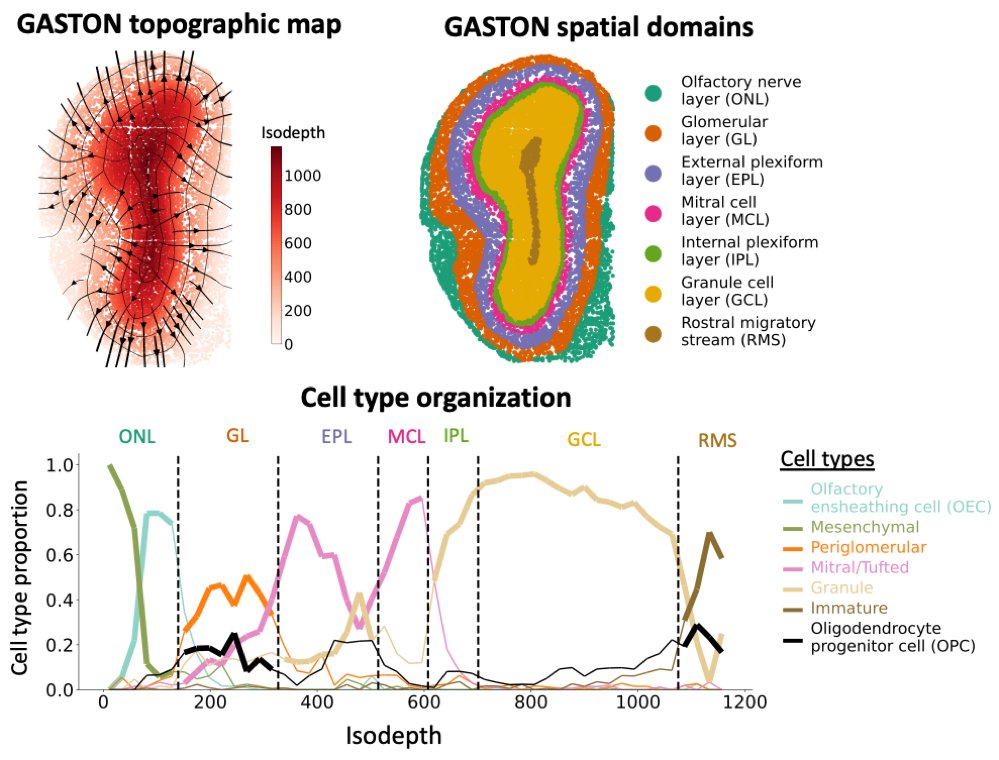
In the olfactory bulb, the GASTON isodepth indicates the “differentiation potential” of neurons – with isodepth gradients showing trajectories of neural maturation. Moreover, GASTON reveals the distinct layers and cell type gradients within the layers. (6/7)
1
0
6
GASTON also identifies gradients in the tumor microenvironment; e.g. gradients of metabolic activity in the tumor interior, and gradients of EMT-related genes in the adjacent stroma – indicating possible metastatic migrations. (5/7)
1
0
3
By aggregating sparse gene expression measurements across contours of equal isodepth, GASTON identifies expression gradients in complex tissue geometries, e.g. the curved layers of the cerebellum. (4/7)
1
0
3
To derive such a topographic map, we introduce the *isodepth*: a 1-D coordinate in a tissue slice analogous to elevation in a topographic map. GASTON learns the isodepth using an interpretable deep learning model. (3/7)
1
0
3
From a topographic map of a landscape, a hiker can identify mountains and valleys or determine the steepness of a path. Similarly, from a topographic map of a tissue slice, one can find spatial domains in a tissue and examine gradients of gene expression within domains (2/7)
1
0
3
Our new method GASTON learns a “topographic map” of a tissue slice, enabling simultaneous identification of spatial domains and gene expression gradients in spatial transcriptomics https://t.co/mnZSxzUkqx Led by @UthsavC w/ @BrianJohnArnold @hrksrkr @Cong992 Sereno + Kohei (1/7)
4
61
232
Consider this great opportunity to submit algorithmic or analysis cancer comp bio contributions to present to the phenomenal @RECOMBconf community and to be considered for PLOS CB! Deadline extended but happening soon!
We have extended the RECOMB-CBB deadline for papers submission to February 7, 2023 23:59, AoE. Send your #Cancer #Computational #Biology papers to #RECOMB_CCB at @RECOMBconf. This year's edition in Istanbul follows the previous amazing @2022Ccb ! #rt
https://t.co/0mg3sK4k7P
0
1
1
Belayer first appeared @Recomb2022 and in preprint form ( https://t.co/yYoQrRTHQ2). Excited to be one of several exciting papers in the “Focus on #recomb2022” special section in @CellSystemsCP 6/6
biorxiv.org
Spatially resolved transcriptomics (SRT) technologies measure gene expression at known locations in a tissue slice, enabling the identification of spatially varying genes or cell types. Current...
0
0
0
Belayer software and tutorial available on Github: https://t.co/Z5gNd4UEBA 5/6
github.com
Contribute to raphael-group/belayer development by creating an account on GitHub.
1
1
1
By using a global model of tissue organization, Belayer accurately identifies tissue layers and spatially varying genes in the brain and skin. 4/6
1
0
0
What about tissues with curved layers? Belayer “straightens” the curves using the theory of conformal maps from complex analysis, or in the simpler case by modeling heat diffusion through the layers with the isotherms defining the relative depth within a layer. 3/6
1
0
1
Belayer models gene expression as a piecewise linear function of layer depth – accounting for both continuous gradients of expression within a layer and discontinuities in expression at layer boundaries – and combining information across spots at same depth 2/6
1
0
0
Spatial transcriptomics meets complex analysis – our new method Belayer models spatial gene expression in layered tissues, now published in @CellSystemsCP. Work led by @Cong992 and @uthsavc with Shirley Zhang. 1/6 https://t.co/OV0Fq0Hsyo
2
49
175
Five computer science graduate students have been named 2023 @SiebelScholars. The award recognizes academic achievement and leadership. Congratulations! 🎉 https://t.co/DSHPPAcQ9X
2
3
26
Princeton Bioengineering is hiring! (please ReTweet!). The search is open rank, but we're particularly interested in talented postdocs on the faculty job market. Applications due Nov15. Come join our vibrant and growing Bioengineering community! https://t.co/Y6rfDVHZOG
0
37
53