Roberto Chica Lab
@Chica_lab
Followers
1K
Following
907
Media
48
Statuses
698
Our research group at the University of Ottawa specializes in protein engineering and computational protein design. #proteinengineering #proteindesign
Ottawa, Ontario
Joined November 2017
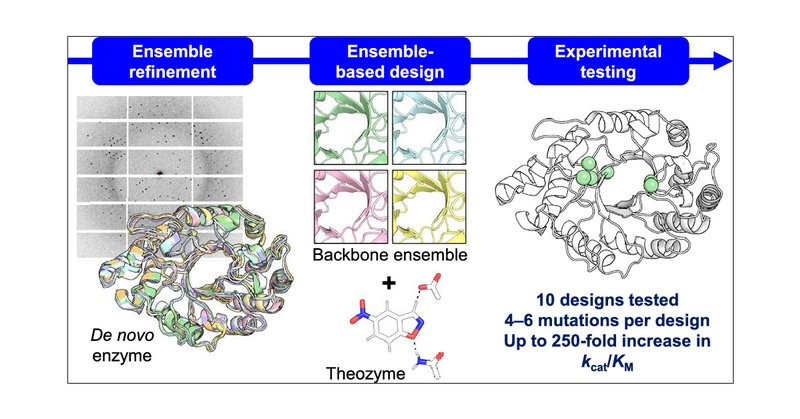
Happy to announce our latest publication on #enzymedesign, in collaboration with @MThompsonLab! Check it out in @J_A_C_S: Design of Efficient Artificial Enzymes Using Crystallographically Enhanced Conformational Sampling
pubs.acs.org
The ability to create efficient artificial enzymes for any chemical reaction is of great interest. Here, we describe a computational design method for increasing the catalytic efficiency of de novo...
2
21
109
I am very excited to share that our paper led by @erdogananisan, @PouriaDasmeh, @AdrianSerohijos is out @natcomms. It touches key dynamics to understand protein evolution"Neutral Drift" and "Phenotypic Variation" during enzyme/protein evolution. https://t.co/EOJTGv8MNc
nature.com
Nature Communications - Threshold-like relationships between fitness & phenotype are ubiquitous in nature. Erdoğan et al. use experimental & simulated evolution of an...
1
11
27
Come see the scientists that should've shared the Nobel Prize for "Computational Protein Design", speak: https://t.co/vQMNKW1xMP
2
90
398
We are recruiting: @ProtEngDesSel is looking for an Editor-in-Chief to lead the journal into the future! If you’re a leader in protein engineering with a strong vision for the field, we want to hear from you. View more details and apply: https://t.co/wqYwxOvFhQ
#JournalEditor
0
2
1
BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to David Baker “for computational protein design” and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction.”
515
9K
20K
For more information, check out the nice editorial written by @ferruz_noelia and @_amelie_rocks: https://t.co/67gIQGBPBl
academic.oup.com
Computational protein design (CPD) aims to create proteins with novel functions or properties not found in nature. While CPD has a long history (Dahiyat an
0
3
8
Happy to announce the special article collection on computational methods for #ProteinDesign, which I edited for @ProtEngDesSel.
@ProtEngDesSel is excited to share its latest article collection on "Computational Methods for #ProteinDesign". Free to read until Sept. 30, 2024:
1
0
11
We’re thrilled to share our new pre-print exploring how we can design and control dynamic proteins! (1/6) https://t.co/w7sRsdJV9y
5
115
505
Today in @Nature we present the results of our exploration of the soluble protein universe and the design of functional soluble membrane proteins with @CasperGoverde @GoldbachNico @befcorreia 1/6 https://t.co/Dakeh8Hyw7
21
206
872
We investigated AlphaFold3 against RoseTTAfold-AllAtom and a new variant of Metal3D for prediction of metal-protein complexes. Tl;dr AlphaFold3 is as good as specialized tools on metal ion prediction. RFAA not so much. Some interesting findings in below thread: 1/
3
43
222
An interesting nugget in the latest paper from @MoAlQuraishi lab: AFDB models are less designable in ways that suggest inaccuracies at the local level but not global level https://t.co/9sX2IVjhyr
5
6
66
This is a complex discussion that I am hoping will continue well beyond this letter. We need to find standards that work in an increasingly complex landscape of research and publishing, and that balances diverse interests. We need to find solutions that enable science to ...
We were incredibly disappointed with the lack of code or executable accompanying the publication of AF3 in @Nature . This contradicts scientific principles of the ability to evaluate, use, and build upon existing work.
1
16
58
Join us! We are looking for a new team member (PhD student) with strong background in organic chemistry. Organic chemistry meets #DirectedEvolution Highly interdisciplinary & passionate research group 🙏RETWEET (We want to recruit internationally) https://t.co/xol9YRph9u
0
31
73
Design of Efficient Artificial Enzymes Using Crystallographically Enhanced Conformational Sampling | Journal of the American Chemical Society @ucmerced @uOttawa @Chica_lab @uOttawaScience #Artificial #Enzyme #Crystallographic #Conformational #Sampling
pubs.acs.org
The ability to create efficient artificial enzymes for any chemical reaction is of great interest. Here, we describe a computational design method for increasing the catalytic efficiency of de novo...
0
4
19
Design of Efficient Artificial Enzymes Using Crystallographically Enhanced Conformational Sampling
pubmed.ncbi.nlm.nih.gov
The ability to create efficient artificial enzymes for any chemical reaction is of great interest. Here, we describe a computational design method for increasing the catalytic efficiency of de novo...
0
1
0
New article from our "Computational methods for protein design" special collection! By @CompBioLab_IQCC (@IQCCUdG). The shortest path method (SPM) webserver for computational enzyme design
academic.oup.com
Abstract. SPMweb is the online webserver of the Shortest Path Map (SPM) tool for identifying the key conformationally-relevant positions of a given enzyme
0
13
57
The 5th Protein Engineering Canada Conference will take place at the University of Toronto on June 26-28, 2024. Speakers include @ltchong @helloanum @chang_c_liu @DavidKwanPhD @casarkar @wenjingwanglab @WeiZhangTO and more! https://t.co/Mcg4RMv4CC
0
8
22