PEDS: Protein Engineering, Design & Selection
@ProtEngDesSel
Followers
1K
Following
32
Media
15
Statuses
230
PEDS is an Oxford University Press journal publishing the latest research in #proteinengineering, #proteindesign and #proteinscience.
Joined January 2020
@ProtEngDesSel is excited to share its latest article collection on "Computational Methods for #ProteinDesign". Free to read until Sept. 30, 2024:
academic.oup.com
Protein Engineering, Design and Selection is pleased to publish a special collection of articles on computational methods for protein design. Computational
1
10
28
We are recruiting: @ProtEngDesSel is looking for an Editor-in-Chief to lead the journal into the future! If you’re a leader in protein engineering with a strong vision for the field, we want to hear from you. View more details and apply: https://t.co/wqYwxOvFhQ
#JournalEditor
0
2
1
Congratulations to @NobelPrize winner David Baker for his groundbreaking contributions to protein design! Learn more about his research in the webinar linked below: https://t.co/jzoixybvZB.
BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to David Baker “for computational protein design” and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction.”
0
2
4
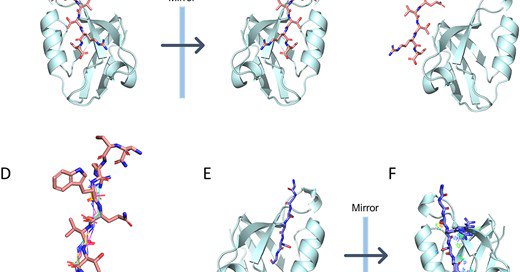
10/ Bruce Donald and coworkers (@dukecompsci) introduce DexDesign, an extension of their OSPREY #ProteinDesign software that enables the creation of D-peptide inhibitors. https://t.co/hYSCfDKNRh
academic.oup.com
Abstract. With over 270 unique occurrences in the human genome, peptide-recognizing PDZ domains play a central role in modulating polarization, signaling,
0
0
0
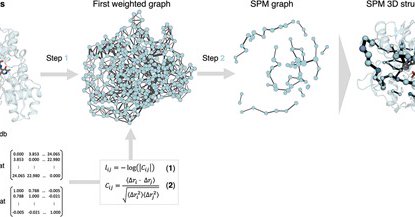
9/ @GuillemCasaF and @silviaosu from @CompBioLab_IQCC (@IQCCUdG) present a web server for the computation of shortest path maps for any enzyme of interest. https://t.co/GAOT3wfmZu
academic.oup.com
Abstract. SPMweb is the online webserver of the Shortest Path Map (SPM) tool for identifying the key conformationally-relevant positions of a given enzyme
1
0
2
8/ @ProteinMagnus and coworkers (@MolBiolAU) review the design of functional intrinsically disordered proteins (IDPs). https://t.co/sIazaG00ud
academic.oup.com
Abstract. Many proteins do not fold into a fixed three-dimensional structure, but rather function in a highly disordered state. These intrinsically disorde
1
0
0
7/ @ChrisWellsWood and colleagues (@SBSatEd) report the Three-dimensional Inference Method for Efficient Design (TIMED), a convolutional #NeuralNetwork-based algorithm for #InverseFolding. https://t.co/asXOftjlll
academic.oup.com
Abstract. Sequence design is a crucial step in the process of designing or engineering proteins. Traditionally, physics-based methods have been used to sol
1
1
3
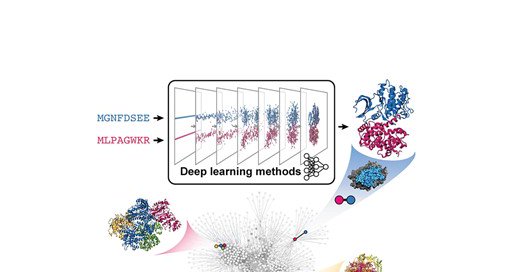
6/ Haiyan Liu and colleagues describe recent advances in #DeepLearning for sequence design on a given protein backbone, also known as #InverseFolding. https://t.co/ftK2XBaAi3
academic.oup.com
Abstract. Deep learning methods for protein sequence design focus on modeling and sampling the many- dimensional distribution of amino acid sequences condi
1
1
2
5/ @JuliaRuRogers, Gergő Nikolényi and @MoAlQuraishi (@ColumbiaSysBio) review the growing ecosystem of deep learning methods for modelling protein–protein interactions. https://t.co/gX8atRQY2p
academic.oup.com
Abstract. Numerous cellular functions rely on protein–protein interactions. Efforts to comprehensively characterize them remain challenged however by the d
1
0
4
4/ Opuu and Simonson (@Polytechnique) review the application of computational design methods to noncanonical amino acids for #GeneticCodeExpansion. https://t.co/THIFZqd16a
academic.oup.com
Abstract. Enzyme design is an important application of computational protein design (CPD). It can benefit enormously from the additional chemistries provid
1
0
1
3/ @KevinKaichuang and colleagues (@MSFTResearch) explore a multimodal approach for inverse folding by training a masked inverse folding protein language model parameterised as a structured graph neural network: https://t.co/RF7ExpaBSQ
academic.oup.com
Abstract. Self-supervised pretraining on protein sequences has led to state-of-the art performance on protein function and fitness prediction. However, seq
1
2
10
2/ Lee et al. (@protabit) present the Triad Antibody Homology Modelling (TriadAb) package, which predicts the structure of heavy and light chain sequences of an #antibody Fv domain using template-based modelling. https://t.co/YT1xBjhs6g
academic.oup.com
Abstract. Computational modeling and design of antibodies has become an integral part of today’s research and development in antibody therapeutics. Here we
1
0
0
@ferruz_noelia @_amelie_rocks 1/ @BirteHoecker et al. use a physics-based approach to insert structural elements on a de-novo-designed TIM barrel: Physics-based approach to extend a de novo TIM barrel with rationally designed helix-loop-helix motifs https://t.co/tiGpsPqklH
academic.oup.com
Abstract. Computational protein design promises the ability to build tailor-made proteins de novo. While a range of de novo proteins have been constructed
1
2
5
This collection provides a broad view of the current diversity and progress within the field of #ProteinDesign, spanning classic modelling approaches and newer machine learning methods. Check out the editorial by @ferruz_noelia and @_amelie_rocks:
academic.oup.com
Computational protein design (CPD) aims to create proteins with novel functions or properties not found in nature. While CPD has a long history (Dahiyat an
1
5
8
Computational methods for protein design
academic.oup.com
Computational protein design (CPD) aims to create proteins with novel functions or properties not found in nature. While CPD has a long history (Dahiyat an
0
28
80
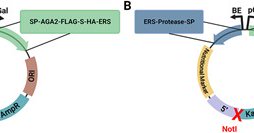
New article alert! @CarlDenard et al. present a series of YESS plasmids that provide a robust platform to observe and quantify PTM-enzyme activity in yeast. Modular and integrative activity reporters enhance biochemical studies in the yeast ER
academic.oup.com
Abstract. The yeast endoplasmic reticulum sequestration and screening (YESS) system is a broadly applicable platform to perform high-throughput biochemical
0
3
3
Check our latest work about shortest path method (SPM) webserver for computational enzyme design by @GuillemCasaF J Casadevall @CrisDuran25 @silviaosu in @ProtEngDesSel
#IQCCpaper @TCBioSys @UdGRecerca
https://t.co/3J5Xr7YRX2
academic.oup.com
Abstract. SPMweb is the online webserver of the Shortest Path Map (SPM) tool for identifying the key conformationally-relevant positions of a given enzyme
0
3
17
New article alert! @CarlDenard et al. present a series of YESS plasmids that provide a robust platform to observe and quantify PTM-enzyme activity in yeast. Modular and integrative activity reporters enhance biochemical studies in the yeast ER
academic.oup.com
Abstract. The yeast endoplasmic reticulum sequestration and screening (YESS) system is a broadly applicable platform to perform high-throughput biochemical
0
3
3
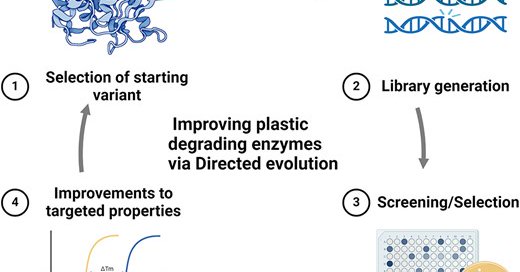
New article alert! In this review, @Jackson_Lab identify knowledge gaps and current challenges, and highlight recent studies related to the directed evolution of plastic-degrading enzymes. Improving plastic degrading enzymes via directed evolution
academic.oup.com
Abstract. Plastic degrading enzymes have immense potential for use in industrial applications. Protein engineering efforts over the last decade have result
0
3
15
New article from our "Computational methods for protein design" special collection! DexDesign: an OSPREY-based algorithm for designing de novo D-peptide inhibitors
academic.oup.com
Abstract. With over 270 unique occurrences in the human genome, peptide-recognizing PDZ domains play a central role in modulating polarization, signaling,
0
1
12