Yuji Sakai (境祐二)
@yj_sakai
Followers
75
Following
210
Media
0
Statuses
64
I work on theoretical studies of intracellular organization. chromosome formation, membrane morphogenesis, atomic and/or coarse-grained simulations
Joined October 2021
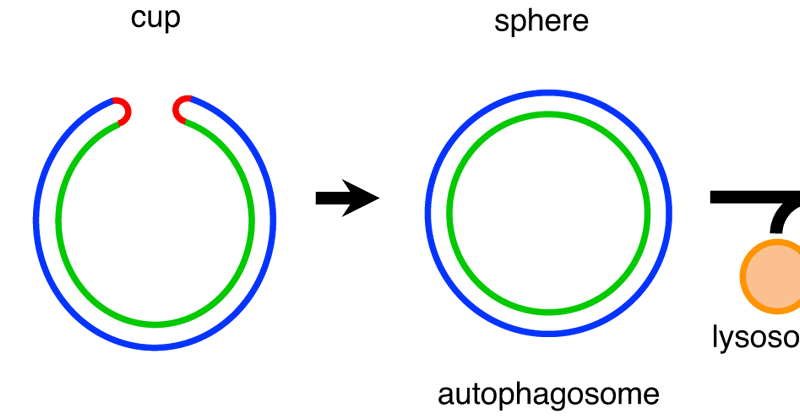
We published the paper "Experimental Determination and Mathematical Modeling of the Standard Shapes Forming Autophagosomes" in Nat. Commun.
nature.com
Nature Communications - Autophagosome formation involves membrane morphological changes. Here, authors statistically determined average shapes of forming autophagosomes from 3D electron micrographs...
1
2
11
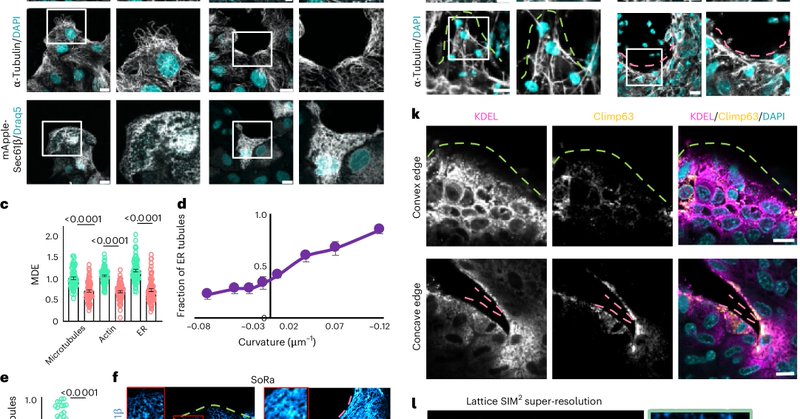
RT @cell_club: Edge curvature drives endoplasmic reticulum reorganization and dictates epithelial migration mode.
nature.com
Nature Cell Biology - Rawal et al. combine intracellular cartography and biophysical modelling to reveal how edge curvature governs endoplasmic reticulum (ER) morphology, showing that...
0
3
0
RT @biorxiv_biophys: The molecular mechanism of lipid uptake by membrane-anchored bridge-like lipid transfer proteins. .
biorxiv.org
Lipid transport by bridge-like lipid transfer proteins (BLTPs) is emerging as a key process in lipid and cellular metabolism in both physiological and pathological conditions. However, the precise...
0
5
0
RT @biorxiv_cellbio: Architecture and Function of Holocentric CENP-A-Independent Kinetochores #biorxiv_cellbio.
biorxiv.org
Kinetochores are essential macromolecular complexes that anchor chromosomes to the mitotic spindle to ensure faithful cell division[1][1]. Despite their critical role, the structural organization of...
0
3
0
RT @CG_Martini: Nice work from Chelsea Brown: she used integrative modelling to build and simulate a mitochondrial cristae (.
0
15
0
RT @biorxiv_biophys: Helfrich Monte Carlo Flexible Fitting: physics-based, data-driven cell-scale simulations #bior….
biorxiv.org
Simulating entire cells represents the next frontier of computational biology. Achieving this goal requires methods that accurately reconstruct and simulate cellular membranes across spatial and...
0
2
0
Excited to share our #biorxiv preprint.
Mechanism of bridge-type phospholipid transfer by Atg2 for autophagosome biogenesis #biorxiv_cellbio.
1
1
7
RT @biorxiv_cellbio: TMEM170 family proteins are lipid scramblases that physically associate with bridge lipid transporters BLTP1/Csf1 http….
biorxiv.org
Bulk lipid transport between organelles has been proposed to involve the partnership between bridge lipid transport proteins and membrane-embedded lipid scramblases. However, for almost all BLTPs,...
0
4
0
RT @PhysRevResearch: How cells wrap around coronavirus-like particles using extracellular filamentous protein structures, Sarthak Gupta et….
0
5
0
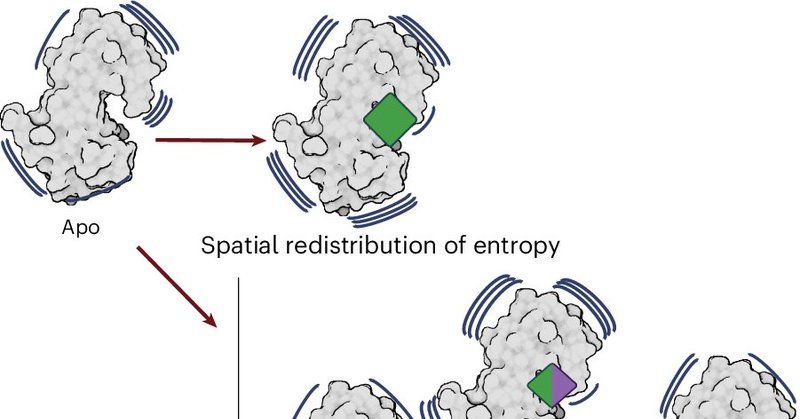
RT @nchembio: A Perspective by Stephanie Wankowicz and James Fraser discusses ways macromolecules use conformational entropy to control bin….
nature.com
Nature Chemical Biology - Protein conformational entropy plays a vital role in functions like binding and catalysis. This Perspective discusses three ways macromolecules use conformational entropy:...
0
26
0
RT @JohnFMarko: Subchromosomal mechanics of pericentric and centromeric chromatin along stretched mitotic chromosomes - surprise is little….
0
6
0
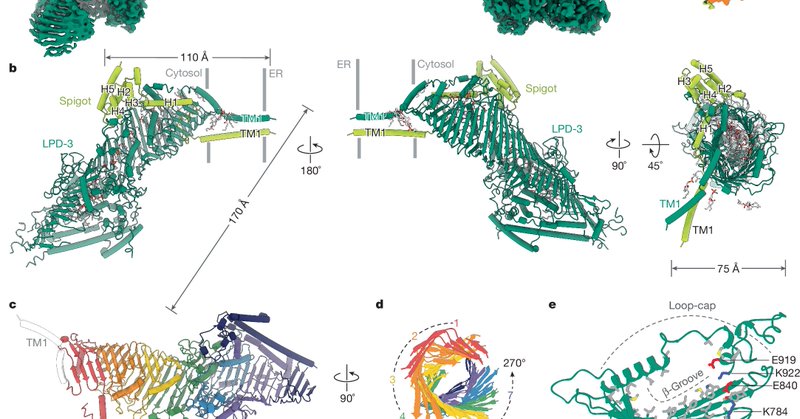
RT @Nature: Nature research paper: Structural basis of lipid transfer by a bridge-like lipid-transfer protein.
nature.com
Nature - The LPD-3 complex structure reveals protein–lipid interactions that suggest a model for how the native LPD-3 complex mediates bulk lipid transport and provides a foundation for...
0
35
0
RT @WPezeshkian: New preprint: TS2CG v2, building and simulating membranes with any shape and lateral organisation. We got Martini Globe, M….
0
14
0
RT @SergioCruzLeon1: Excited to share our pre-print on the molecular architecture of heterochromatin in human cells🧬🔬.w/ @jpkreysing, Johan….
0
120
0
RT @CellCellPress: Now online! Cell membranes sustain phospholipid imbalance via cholesterol asymmetry https://t.c….
0
60
0
RT @ScienceMagazine: In a new Science study, cryo–electron tomography captures the in-cell architecture of the mitochondrial respiratory ch….
0
146
0
RT @yiechanglin: 1/ In my final project as part of the @CorryLab, we show the bacterial TAM complex facilitates the spontaneous flow of pho….
0
33
0
RT @BiologyAIDaily: Machine Learning Methods to Study Sequence–Ensemble–Function Relationships in Disordered Proteins . 1/ This review exp….
0
5
0
RT @Austin_Lefebvre: Do you LOVE organelles??.Well I am THRILLED to announce that Nellie has been published in @naturemethods! . Nellie a f….
0
114
0