Austin E. Y. T. Lefebvre
@Austin_Lefebvre
Followers
886
Following
9K
Media
42
Statuses
539
Dr. Leafbeaver, DS @calico. You can't spell imaging without "im aging" and boy am I. Organelle enthusiast. Creator of Nellie and Mitometer.
San Francisco, CA
Joined September 2012
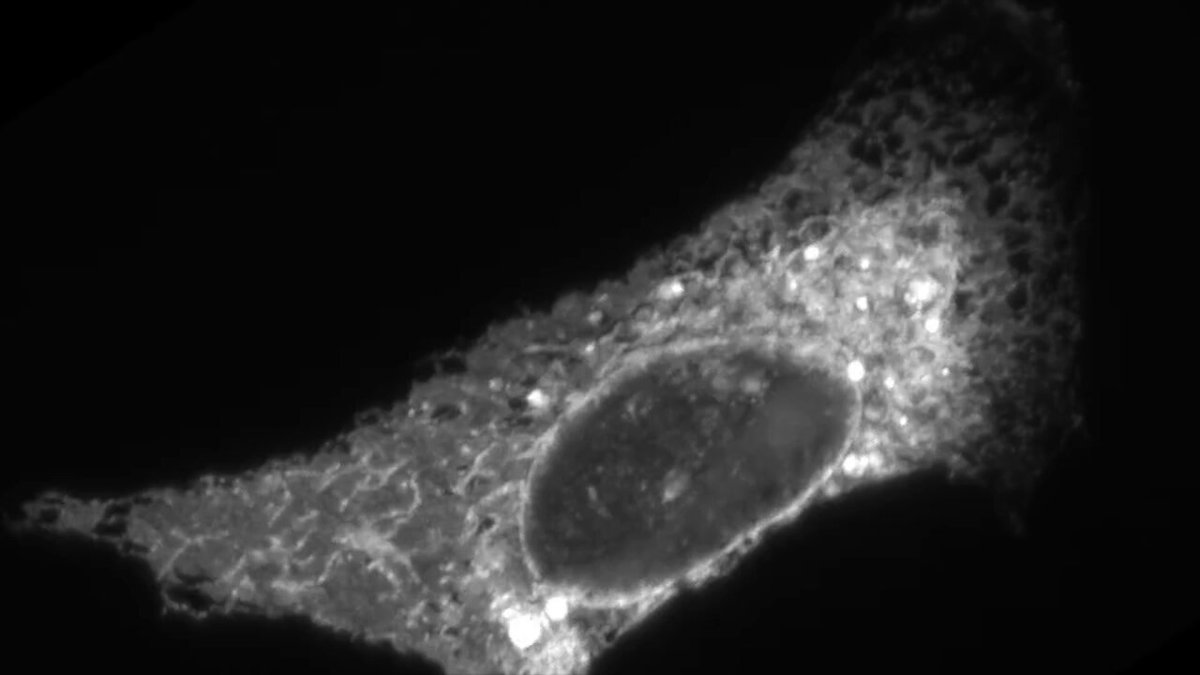
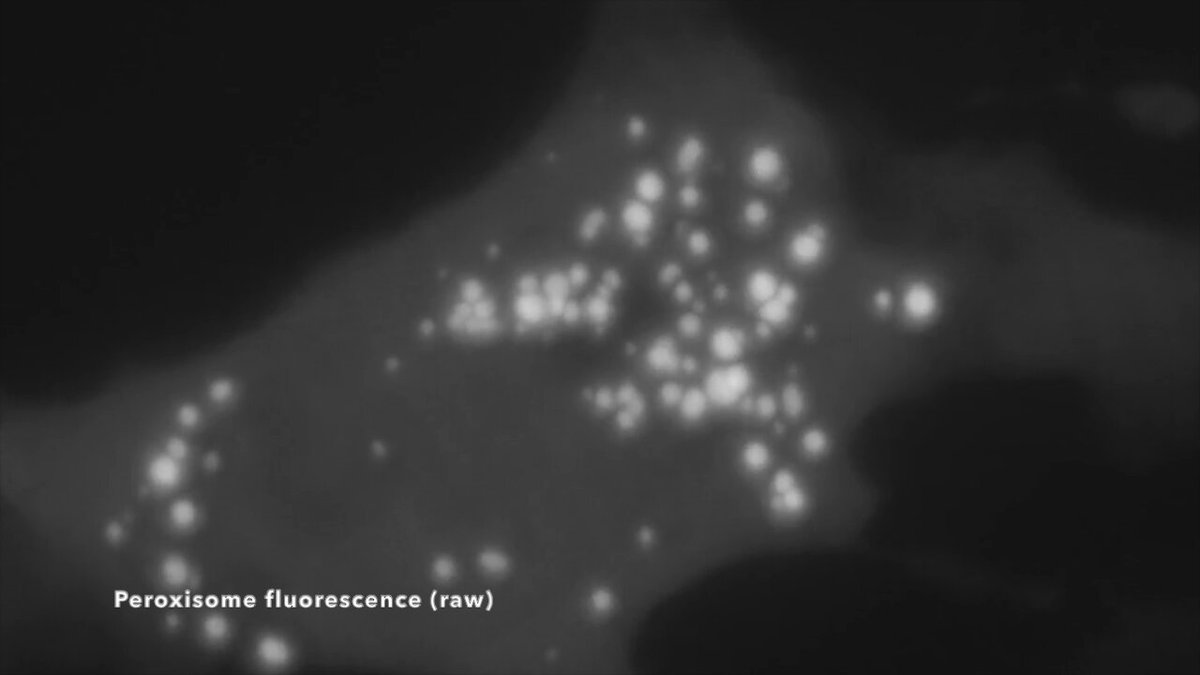
Do you LOVE organelles??.Well I am THRILLED to announce that Nellie has been published in @naturemethods! . Nellie a fully automated pipeline for organelle segmentation, tracking, and hierarchical feature extraction in 2D, 3D, timelapse, multichannel live-cell microscopy. 🧵1/N
12
114
507
RT @manorlaboratory: My lab’s 5-year NIH R01 grant, awarded to study gene therapy for hearing loss, was abruptly terminated. I want to shar….
0
2K
0
RT @Austin_Lefebvre: Analyzing organelles is HARD. They're complex, but critical to understanding biology. Current tools are manual, organe….
0
3
0
RT @t_andy_keller: In the physical world, almost all information is transmitted through traveling waves -- why should it be any different i….
0
927
0
Already some cool applications for Nellie: If you're used to MitoGraph data and want some of the same outputs from Nellie, check out Gav's Python code.
Nellie's image analysis capabilities are vast. I put a video of yeast mitochondria through Nellie and generated a 'live-cell MitoGraph' - a topological graph capturing mitochondrial network's fission and fusion events over time.
0
3
12
RT @naturemethods: New from Lefebvre and colleagues! Nellie is a comprehensive automated pipeline for studying the structure and intracellu….
0
8
0
Before deep-diving into Nellie, shout out my co-authors: @sturm_gav, for help on all the paper, experiments, and testing, and to the other co-authors @Kayley_Hake, @bbkaufmann @emistoops, Magdalena Lopez, and Molly Lin. And of course @rita_strack for being a great editor!. 🧵8/N.
1
0
4
And just for fun, here's me walking around, masked by Meta's Segment Anything Model (@AIatMeta), and tracked via Nellie's tracking pipeline. 🧵7/N
1
0
4
Inspired by @GoogleDeepMind's weather forecasting model GraphCast, we also developed an organelle graph autoencoder using Nellie's outputs to analyze mitochondrial networks' responses to ionomycin treatment, revealing complex dynamics invisible to traditional analysis. 🧵4/N
1
2
10
Nellie is FULLY automated with NO parameters to tune, and works on ALL structures - from mitochondria to ER to microtubules and beyond. (credit to @AndrewGYork and @amsikking's Snouty SOLS microscope for the data, and @AlisterBurt for the Napari Animation plugin). 🧵3/N
1
0
15