Teeradon Phlairaharn
@teerap16
Followers
249
Following
7K
Media
29
Statuses
444
Student @tu_muenchen Journey through @labs_mann | @erwinschoof | @briansearle | @jesperolsenlab | @hamish_stewart
Munich, Bavaria
Joined July 2019
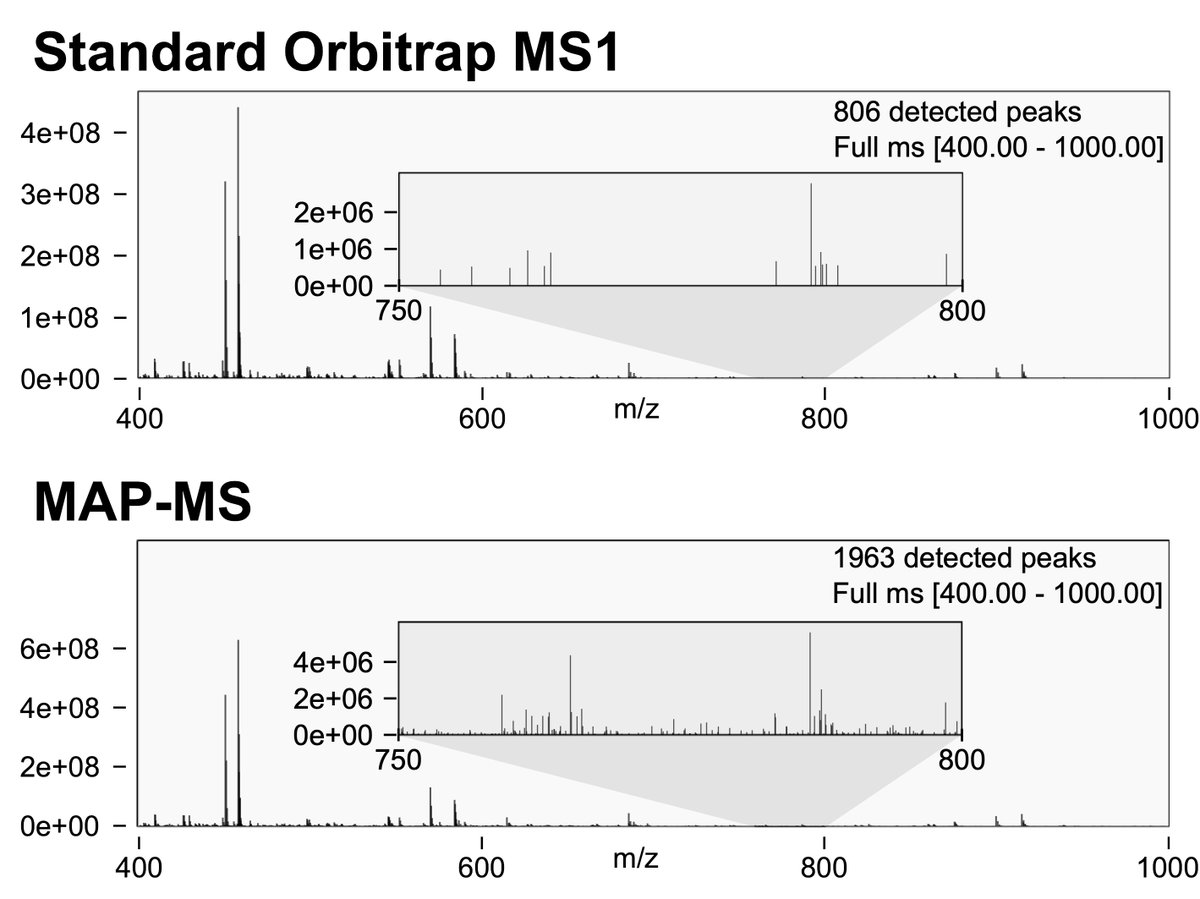
Give MAP-MS a try on your Orbitrap- no hardware or software changes needed! Curious how it works? Check out @briansearle tweet for the details. #MassSpec #Orbitrap #asms2025 Huge thanks to coauthors and everyone who supported us during this work!.
People have asked about how MAP-MS works so here's a tweetorial on @teerap16's new preprint: MAP-MS takes advantage of parallelization in a trapping MS, collecting Orbitrap MS1s with 2x the dynamic range by differentially amplifying parts of the ion beam.
0
7
13
RT @biorxiv_biochem: Improving proteomic dynamic range with Multiple Accumulation Precursor Mass Spectrometry (MAP-MS) .
biorxiv.org
Orbitrap (OT) -based mass spectrometer platforms are a gold standard in high-resolution mass spectrometry, where their primary disadvantage is slower-scanning speed in comparison to time-of-flight or...
0
1
0
RT @Earthnut89: ENVLPE, our system for VLP-based delivery of gene editors, is now published in @CellCellPress. We s….
0
6
0
RT @JesperOlsenLab: 🚨 Our #singlecell article is out! .We describe the Chip-Tip label-free SCP workflow quantifying >5000 proteins in singl….
nature.com
Nature Methods - Chip-Tip is a label-free quantification-based single-cell proteomics workflow for deep single-cell proteomics, which identifies over 5,000 proteins and 40,000 peptides in single...
0
9
0
RT @jespervolsen: We are excited to share our new Nature Methods paper describing the Chip-Tip workflow for single-cell proteomics identify….
0
38
0
RT @niggles293T: Excited to share 'non-destructive transcriptomics via vesicular export' (NTVE). We use HIV-1 virus-like particles, to exp….
biorxiv.org
Transcriptomics is a valuable technique for multiplexed monitoring of cellular states. However, its application has been predominantly limited to assays that necessitate cell fixation or lysis, which...
0
2
0
RT @Earthnut89: Check out our approach on "non-destructive transcriptomics via vesicular export" (NTVE)! We have benchmarked NTVE exclusive….
biorxiv.org
Transcriptomics is a valuable technique for multiplexed monitoring of cellular states. However, its application has been predominantly limited to assays that necessitate cell fixation or lysis, which...
0
5
0
It was a pleasure to have a wonderful lunch together during #HUPO2024 @briansearle @wilhelm_compms @Hamish_Stewart .
0
0
10
It's a wonderful opportunity at #HUPO2024 to reconnect with my incredible mentors. @Hamish_Stewart @briansearle 😙
0
1
14
RT @wilhelm_compms: You have a strong background in bioinformatics🖥️, data science📈, machine learning, and multi omics? You want to do a P….
0
15
0
RT @Hamish_Stewart: For those attending the final morning of #IMSC2024, I'm giving a talk covering decades of secret instrument development….
0
3
0
RT @massspecpro: [ASAP] Racetrack FAIMS for High-Resolution and High-Sensitivity Characterization of Peptide Conformers .
pubs.acs.org
A racetrack field asymmetric waveform ion mobility spectrometry (r-FAIMS) device, which consists of both cylindrical FAIMS (c-FAIMS) and planar FAIMS (p-FAIMS) sections with a 1 mm gap width, was...
0
2
0
RT @Eileen_Furlong: A heads up to all post-docs - the Genome Biology dept @EMBL is recruiting - independent group leader (PI) position. Out….
0
110
0
RT @jaybradner: Today in @ScienceMagazine, together with my former colleagues at NIBR, we report the discovery and characterization of firs….
0
134
0
RT @briansearle: 1/8 I’m excited to share our new work by @ArianaShannon on low-input targeted proteomics with a brand-new linear ion trap!….
biorxiv.org
Advances in proteomics and mass spectrometry have enabled the study of limited cell populations, such as single-cell proteomics, where high-mass accuracy instruments are typically required. While...
0
14
0