Anoushka
@noush_joglekar
Followers
732
Following
3K
Media
13
Statuses
705
Tweets about science and the occasional musing Single-cell | Splicing | Epigenetics | Tech dev Previously postdoc @nygenome and student @WeillCornell she/her
New York, NY
Joined November 2017
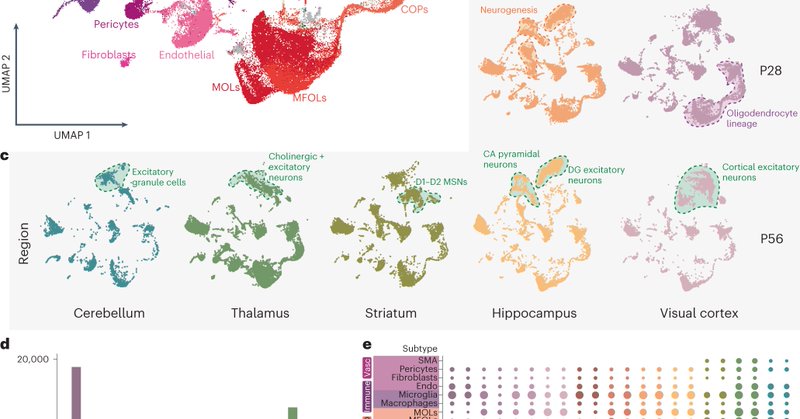
If you were looking for evidence that isoforms are cool, all-pervasive in the 🧠, and everybody should study them at the single cell level, our work on mouse development and brain-region variability from the @hagentilgner lab is now live @NatureNeuro! 1/.
nature.com
Nature Neuroscience - RNA alternative splicing is involved in determining cell identity, but a comprehensive molecular map is missing. Here, the authors provide a human and mouse brain atlas of...
10
45
153
RT @ItaiYanai: When your kids ask you what you did to save US science in the crisis of ‘25, you will say that you showed up! Please spread….
0
146
0
RT @JackHumphrey_: Ecstatic to see our paper "Long-read RNA sequencing atlas of human microglia isoforms elucidates disease-associated gene….
nature.com
Nature Genetics - This isoform-centric microglia genomic atlas includes 35,879 novel human microglia isoforms identified by long-read RNA sequencing. A multi-ancestry quantitative trait locus...
0
34
0
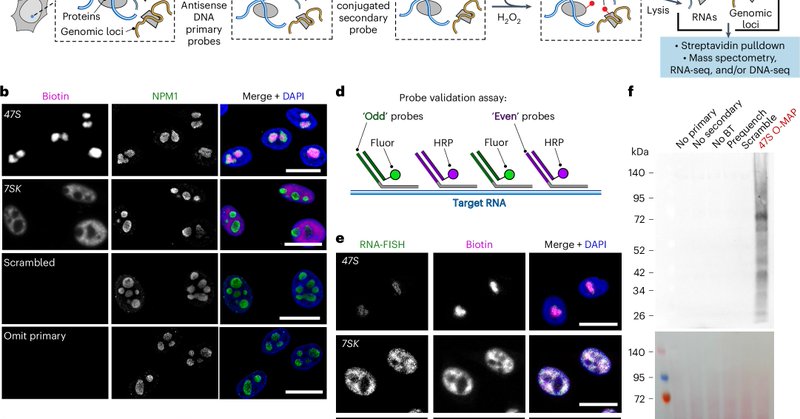
RT @ShechnerLab: Beyond thrilled to present our O-MAP—now *published* @naturemethods (!!!). O-MAP is a powerful new method for biochemicall….
nature.com
Nature Methods - Oligonucleotide-mediated proximity-interactome MAPping (O-MAP) enables precise multiomic characterization of biomolecular interaction networks at target RNAs. Distinct O-MAP...
0
110
0
RT @RongFan8: Today is the day 🤩. Thrilled to share our @Nature paper on Th2 function, CAR-T, & 8-year remission!! Huge team effort w/@Gr….
nature.com
Nature - Elevated type 2 functionality in CAR T cell infusion products is significantly associated with maintenance of a median B cell aplasia duration of 8.4 years in paediatric patients...
0
124
0
RT @amarderstein: 🧵 Thrilled to share our latest in @Nature! We’ve mapped fetal blood development in Down syndrome (DS) using single-cell m….
nature.com
Nature - Using single-cell and multi-omics data of fetal blood, a high-resolution molecular map of dysregulated haematopoiesis in Down syndrome is provided.
0
48
0
RT @ItaiYanai: In today's Cell, we propose a “developmental constraint” model whereby cancer cell state plasticity is restricted by the org….
0
118
0
🤯.
How do enhancers work over distances that sometimes exceed megabases? Excited to share our work led by @gracecbower where we uncover a unique sequence signature globally associated with long-range enhancer-promoter interactions in developing limb buds: 1/
0
0
1
Fantastic first innovation day at London calling @NanoporeConf. I'd say forgetting to tweet = sign of good times.
0
0
6
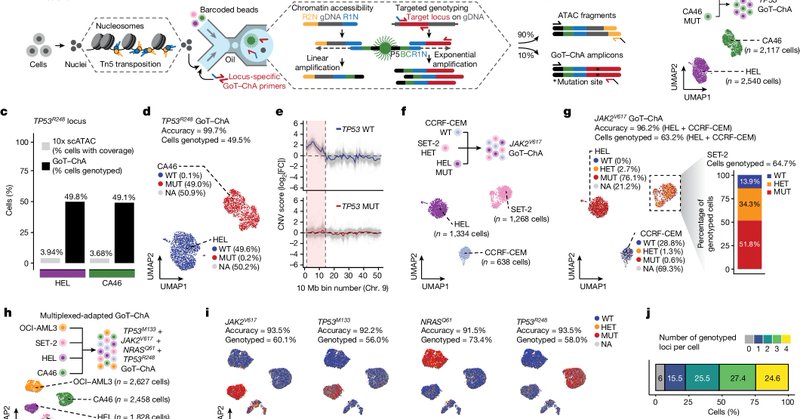
RT @FrancoIzzo85: GoT-ChA! . I am thrilled to announce that our paper "Mapping Genotypes to Chromatin Accessibility Profiles in Single Cell….
nature.com
Nature - The JAK2V617F mutation leads to epigenetic rewiring in a cell-intrinsic and cell-type-specific manner, influencing inflammation states and differentiation trajectories in patients with...
0
99
0
RT @NatureNeuro: Single-cell long-read sequencing-based mapping reveals specialized splicing patterns in developing and adult mouse and hum….
nature.com
Nature Neuroscience - RNA alternative splicing is involved in determining cell identity, but a comprehensive molecular map is missing. Here, the authors provide a human and mouse brain atlas of...
0
32
0
RT @WeillCornell: Researchers use cutting-edge sequencing methods to build the most comprehensive atlas of mRNA variants in the brain. The….
0
6
0
Our data is available already at #NeMO, but we are working on making it widely accessible and interactive - more on that soon! 4/.
0
0
4
Crucial inputs from Betsy Ross, Teri Milner, and @ndhlovulab @WCM_CNG, the team over at @RockefellerUniv @erichjarvis @ofedrigo @JenBalacco @jordanmarrocco, and funding from #BICCN made this study possible 3/.
1
1
3
Loved digging through this data and finding nuggets that eventually made this story. Great teamwork w/ @rainryuhei_wen and fantastic collab w/ @MarkDiekhans @onarykov @DmKorkin @GSheynkman to parse the downstream translational implications and fidelity with human biology 2/.
1
1
4
RT @hagentilgner: Our #BICCN paper is out at @NatureNeuro.Led by @noush_joglekar+@rainryuhei_wen. Single-cell isoforms across brain region….
nature.com
Nature Neuroscience - RNA alternative splicing is involved in determining cell identity, but a comprehensive molecular map is missing. Here, the authors provide a human and mouse brain atlas of...
0
42
0
RT @edeon: Very excited to share our new paper on how mutations in SWI/SNF chromatin remodeling complexes alter chromatin dynamics and immu….
0
65
0
RT @chirag_msk: Thrilled to share our latest work out now in @ScienceMagazine demonstrating a heterozygote advantage at HLA-II in protectin….
0
6
0
Had the best time chatting with @daniel_garalde about the fun I've had looking at single-cell isoforms, and as always at the end of the day there's more questions than answers 🧠.
Ask bolder questions. @noush_joglekar has developed an approach using @10xGenomic libraries & long nanopore reads to investigate isoforms at the single cell level, which may help understand the role they play in different diseases. Learn more: #WYMM
0
1
24