Nathaniel D Anderson
@natedandy_PhD
Followers
184
Following
368
Media
34
Statuses
280
🇬🇧 Staff Scientist at the Sanger Institute | 🇬🇧 Darwin College fellow | 🇪🇺 MSCA alum | 🇨🇦 University of Toronto & SickKids alum
Cambridge, England
Joined January 2016
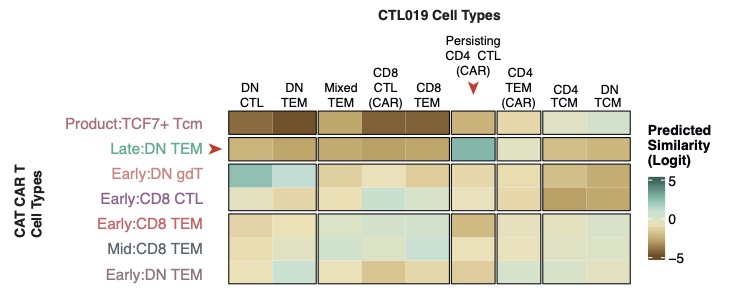
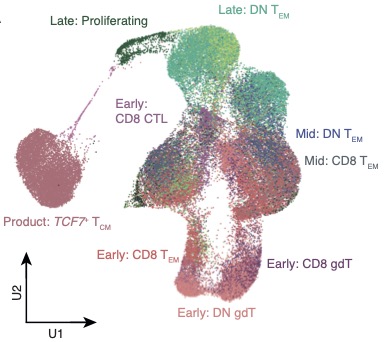
📢 Our paper characterizing long-lived CD19 CAR T-cells from children with B-ALL is out now in @NatureMedicine. 🥳. We performed longitudinal single-cell RNA sequencing of long-lived CAR T-cells and found a transcriptional signature of persistence! .
2
31
101
Amazing day of science and discussion. Thanks to @CLRUKnetwork and the organizers for the opportunity to share our work
0
0
3
Please check out this episode of #EBMT Trainee Pearls where I discuss our recent paper on transcriptional signatures associated with persisting CAR T cells! #CART #leukaemia.
Here's our new episode of Trainee Pearls on transcriptional signatures associated with persisting CD19 CAR-T cells in children with #leukemia with @natedandy_PhD @sangerinstitute in @NatureMedicine . Audio on spotify/apple. #leusm @CarmeloGurnari
0
0
5
RT @sangerinstitute: New research uncovers a genetic signature which explains why some children have a longer remission than others after C….
0
5
0
@PennMedicine And of course thank you to our funding bodies, the patients and their families who make this work possible 🙏. @uclcancer @ucl.@GreatOrmondSt.@sangerinstitute .@wellcometrust .@GOSHCharity .@CwC_UK .@MSCActions.
0
0
0
@PennMedicine THANK YOU to everyone who helped put this study together, especially @sghorashian and Sam. As well as our co-authors (many of which are not on Twitter).@Theo_Accogli .@constantAmateur .@rob_wynn_TdF .@SatyenGohil.
0
0
2
@PennMedicine This MAY suggest that there is a universal transcriptional signature of CAR T persistence that is independent of CAR product, initial cancer type and patient age. (Big maybe at this point).
2
0
1
We describe a persisting CAR T-cell transcriptional signature found in these children that is also observed in two adults with CLL that were treated with a different CAR product @PennMedicine
1
0
1
RT @HuangLabICR: Our new paper "The Proteomic Landscape of Soft Tissue Sarcomas" is out in @NatureComms!. We charac….
0
60
0
I hope everyone is as excited for the @DarwinCollege Lecture Series as I am! @DarwinLectures .
0
0
11
Happy to share our preview on the spatial organization of microbiota in cancer published in @ImmunityCP. Original paper by: .@CJohnstonLab1 @BullmanLab @fredhutch.
3
9
26
RT @EricTopol: Bacteria play a more significant role in cancer than we previously envisioned, as we've recently been learning via spatial g….
0
89
0
RT @mlayeghi: Our new paper is out today on TP53 copy number gain in LFS syndrome! Congrats to @nick_light et al👏👏👏. #ShlienLab #MalkinLab….
0
2
0
Congrats @nick_light et al. on publishing this phenomenal piece of work describing the evolution of Li-Fraumeni tumors. 🎉🎉🎉.
Our study is out today in @NatureComms ! We analyzed cancer genomes from patients with Li-Fraumeni syndrome and found early copy number gain of the mutant allele of TP53 to be a characteristic feature of these tumours.
0
1
7
RT @straathof_karin: 🚨Interested in studying the effect of radiation treatment on the #TME to determine how to combine radio and immune the….
0
14
0