Dan Skelly
@genomequant
Followers
184
Following
356
Media
1
Statuses
46
Data science, systems genetics/genomics, statgen, QTL, single-cell gene expression. I work @jacksonlab but views are my own.
Bar Harbor, ME
Joined October 2017
Our diverse ESC work is out! Amazing collaboration with generous colleagues esp @Laura_Reinholdt C Baker @stevemunger G Churchill + more. https://t.co/kZuhbn2bU8 1/3
New Short Article online now CellStemCell: Mapping the Effects of Genetic Variation on Chromatin State and Gene Expression Reveals Loci That Control Ground State Pluripotency
1
0
6
Interested in doing an genetic experiment with a cutting-edge mouse multiparental population (MPP), but don't know which best suits your needs (e.g., inbred vs outbred)? Using simulations from real genetic data, I sought to provide some answers and guidelines.
Which mouse multiparental population is right for your study? The Collaborative Cross inbred strains, their F1 hybrids, or the Diversity Outbred population https://t.co/kr2RUPkEdf
#bioRxiv
1
10
32
BREAKING via Washington Post: The FBI found documents in Trump's bedroom, office, and the first-floor storage room at Mar-a-Lago during the Aug. 8 search.
410
3K
10K
Genome Sciences mourns the loss of our colleague, Dr. Deborah Nickerson. https://t.co/Eo0XNgMWTw
0
55
199
See our latest officially out in circulation now!
Single-cell transcriptomics identifies two previously undescribed cardiac fibroblast populations that are key drivers of cardiac fibrosis https://t.co/d8CzDBpMNV
#AHAJournals @Alex_R_Pinto @genomequant @pinto_lab @BakerResearchAu @jacksonlab @PAM_latrobe
2
6
29
GBRS manuscript is up on bioRxiv. We show how we apply an allele-specific expression (ASE) algorithm to reconstructing individual imputed genomes/transcriptome, and then, to quantifying gene expression.
Genotype-free individual genome reconstruction of Multiparental Population Models by RNA sequencing data https://t.co/8k6mAlokSd
#bioRxiv
3
16
36
My very favorite visualization -- RNA-Seq of 185 genetically diverse ESC lines: pluripotency genes = highly expressed (as expected) lineage markers = lowly expressed … but check out that variation 🤩! 3/3
0
0
4
@selcan_t has a great explainer that weaves together this paper with our companion work led by @OrtmannDaniel and @VallierLab
https://t.co/IQ0Vl4FxAv 2/3
1
0
1
Really fun collab. I think in the future we will see much more integrating #singlecell across genetic backgrounds ... Congrats @ElviraForte1!
Our paper is officially out! @CellReports Dynamic Interstitial Cell Response during Myocardial Infarction Predicts Resilience to Rupture in Genetically Diverse Mice https://t.co/VKR7HUFuTU
@jacksonlab,@genomequant, @nadia_rosenthal, @vivekepsilon, #SingleCell
1
0
7
very nice paper showing multiple tissue integration of gene expr/ATAC QTL with identification of candidate causal models. Congrats!
Excited to see (and share) the final version of this work with @bryancquach! From the labs of @WilliamValdar, Ivan Rusyn, and Terry Furey. #PLOSGenetics: Integrative QTL analysis of gene expression and chromatin accessibility identifies multi-tissu ...
0
1
9
New tools for integrating GWAS and scRNA-Seq. Really looking forward to diving in and giving them a try!
THRILLED to share our two NEW computational toolkits called CELLEX and CELLECT for integrating GWAS and scRNA-seq to prioritize etiologic cell-types underlying traits and diseases. Tweetorial 1/12 https://t.co/zL3Xt6lpDW
0
0
5
Really interesting paper on microbiome genetics in diverse mice. Congrats @ElissaJChesler @jbubier!
0
3
9
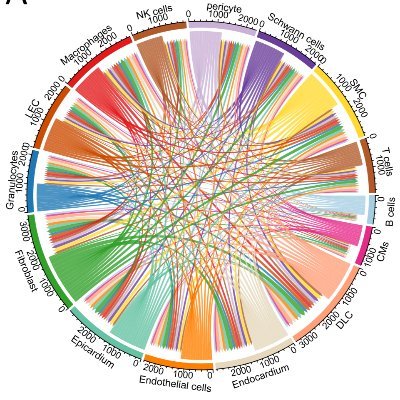
Our attempt at scRNA-Seq profiling of the heart maximizing both data quality and cellular diversity. Homeostasis and AngII model. Really fun study and collaboration between nearly opposite sides of the globe!
Our latest work to help better understand cellular drivers of cardiac fibrosis and hypertrophy. A fantastic team effort by an excellent group of people from @BakerResearchAu @PAM_latrobe @jacksonlab @genomequant. https://t.co/ZgjLLgfKDR
0
3
12
Congratulations @kwangbom ... This work is a tour de force and worth a read for anyone interested in ASE!
[1/n] Multiple bulk RNA-seq studies showed that allelic imbalance (AI) is pretty common. It is subtle in many genes, but sometimes it is extreme as in imprinted or X-inactivated genes. We are curious to see what AI would look like at single-cell resolution.
0
1
5
TT requirement arguably has merit as a shortcut to filtering out unqualified PIs, but I'd push CZI to take the risk of a few unqualified apps in return for engaging with a pool of dynamic pre-TT applicants (assess qualifications using preprints, github, etc). 4/4
1
0
2
Non-TT can be doing great science in an independent capacity (e.g. advanced postdoc or research scientist), may have awesome ideas but lack the resources to bring them to fruition. 3/4
1
0
0
Single cell functional genomics is a rapidly developing field and young people are contributing a lot, including many without TT positions. 2/4
1
0
1
Dear @cziscience: love the new #inflammation in disease https://t.co/PgKOV49X6t and other RFAs. But I have to gripe about eligibility criteria (only PIs with TT position). 1/4
The role of #inflammation in health and disease is not well understood. @ChanZuckerberg’s new call for applications will support science teams to collaborate + apply new tools to gain greater insight into inflammatory diseases
1
2
3
Finally, a principled way to choose how many PCs to use in scRNA-Seq. Looking forward to diving in to the code and trying this on some of my own data!
Single-Cell RNA-seq data is notoriously noisy. There are dozens of methods for cleaning it, but how do you know when they’re working right? How clean is too clean? Presenting...Molecular Cross-Validation https://t.co/5Qeu7jxo4i
1
0
4