David Li
@_David_Li
Followers
804
Following
6K
Media
11
Statuses
892
BioE PhD Student @Stanford with Hie/Fischbach/Deisseroth labs, previously Scheres group @MRC_LMB via @MarshallScholar, Zhang lab @broadinstitute, @MIT '22
Joined April 2018
Had fun solving the cryo structure of Evo-Φ36 with the awesome team of @samuelhking, @driscoll_cl, and @maxewilkinson! Samuel is a beast and the reason there is a 'skevophage' react on Stanford slack.
Many of the most complex and useful functions in biology emerge at the scale of whole genomes. Today, we share our preprint “Generative design of novel bacteriophages with genome language models”, where we validate the first, functional AI-generated genomes 🧵
1
7
87
Hot off the press! Our latest paper led by @fernpizza , understanding how plasmids evolve inside individual cells. These small pieces of self-replicating DNA live inside bacteria and can transfer antibiotic resistance genes, but they also compete with one another to replicate. 1/
6
41
144
🚀New preprint from our lab! I am very excited to finally share what has been the main focus of my PhD for the past almost 3 years at @ETH_en! It is about viral dark matter and a powerful tool we built to shed light on it. 🧬💡 Continue reading (🧵)
3
13
37
Excited to present the first major work after starting our lab at Stanford and the Arc this year: CRISPR-All, a unified genetic perturbation language for programming any major type of genetic perturbation simultaneously, in any combination, at genome scale, in human cells.
8
98
389
🚨 New Results Alert 🚨 Have you ever wondered what new therapeutic avenues would be enabled if we could program T Cells in the computer to target any desired peptide-MHC neoantigen with a TCR or a TCR-like antibody? What if we could validate these molecules easily in small
4
15
52
What is the best strategy to win any contest? Eliminate your opponents of course. Recently, my colleague @fernpizza showed how plasmids compete intracellularly (check out his paper just published in Science today!). Together with @baym, we now know how they can fight.
4
28
78
Today, we present a step-change in robotic AI @sundayrobotics. Introducing ACT-1: A frontier robot foundation model trained on zero robot data. - Ultra long-horizon tasks - Zero-shot generalization - Advanced dexterity 🧵->
423
646
5K
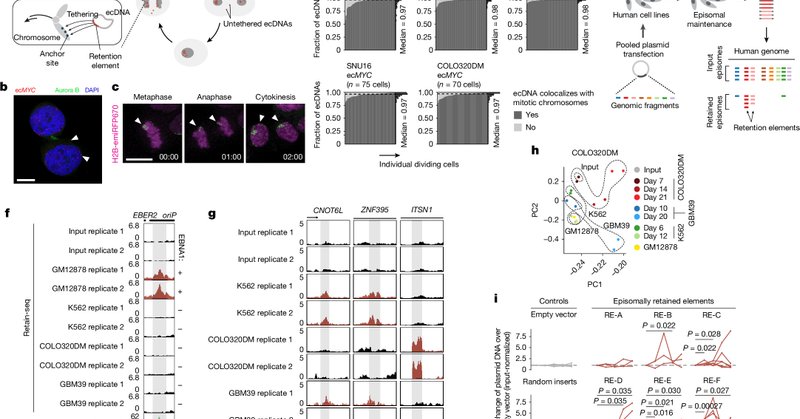
Our paper is out in @Nature! Oncogenes are often copy-number amplified on extrachromosomal DNA (ecDNA) in cancer, but how is ecDNA inherited by dividing cells? Here we identified elements within ecDNA that promote its retention in dividing cells. 1/11 https://t.co/jYAZwj64am
nature.com
Nature - A combination of genome-wide functional screening, imaging and chromatin profiling identifies a new class of highly prevalent genomic elements that help retain extrachromosomal DNA copies...
4
58
252
Today in @Nature we report a new prime editing strategy that can rescue a common cause of many genetic diseases in a disease-agnostic manner. This approach converts a redundant endogenous tRNA into an optimized suppressor tRNA, enabling a single prime edit to rescue premature
32
374
1K
Our new preprint is online! Viruses, bacteria and parasites use effector proteins to evade immunity and rewire host cell pathways. Together with @AlexanderStark8, we wondered if we could systematically map what these effectors, regardless of their origin, do in human cells. 1/8
3
11
49
Major new direction in the @stark_lab: hacking human cell biology with pathogen effectors (eORFs) - amazing collaboration with @mike_tilapia lab. Huge congrats to first authors Tomas & He & all co-authors! Check out the pre-print on @biorxivpreprint
biorxiv.org
Pathogens deploy effector proteins to exploit host cell biology, and most pathogen open reading frames (ORFs) are rapidly evolving and lack functional annotation. We developed the eORFeome, a...
0
17
79
Check out this amazing work by the incredible @aditimerch and team!! Prompting DNA language models a la guilt-by-association lets you design things that function with low seq id
What if we could autocomplete DNA based on function? Today in @Nature, we share semantic design—a strategy for function-guided design with genomic language models that leverages genomic context to create de novo genes with desired functions.🧵 https://t.co/P5qVJB3qIY
0
2
12
I’m really excited about our release of Gemini 3 today, the result of hard work by many, many people in the Gemini team and all across Google! 🎊 We’ve built many exciting new product experiences with it, as you’ll see today and in the coming weeks and months. You can find it
213
357
3K
Check out MitoScribe, our new preprint led by @Linhan_W: https://t.co/rvNOSaMily... It's an analog molecular recorder that uses neutral base edits to the mitochondrial genome to store info about historical signaling in a cell. Single cell resolution at scale (see next post)!
4
8
36
I am excited to announce what I’ve been working on for the past 2 years. General Control is a mandate to develop programmable therapies that make durable, reversible adjustments to gene expression - epigenetically activating or silencing multiple targets at once. In the past 16
93
153
1K
Thrilled to share that I have joined @AnthropicAI as a life science researcher! I am confident that Claude will do amazing things to accelerate biology. Big things ahead!
114
54
2K
#PR Our Ms on brain atlas of circadian activity is out in Science! ~80%(508 regions) of whole brain show significant oscillations. We envisioned it at first CUBIC papers in 2014. It took a decade. Congrats, Katsu&Fukuaki! Ms→ https://t.co/VZNUIEsHbm DB→ https://t.co/01evod4ChG
5
56
212
What if we could watch AND control thousands of individual neurons in behaving mice—with stable access over months? 🧠⚡ Our new preprint introduces transgenic all-optical tools that make this possible - and a resulting surprising discovery! 🔭💡 https://t.co/IvHWooqUmu 1/13 🧵
biorxiv.org
The complexity of the mammalian brain’s vast population of interconnected neurons poses a formidable challenge to elucidate its underlying mechanisms of coordination and computation. A key step...
6
28
133
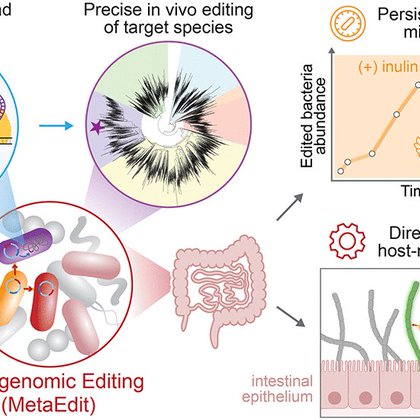
Very excited to share our latest work in Science on metagenomic editing (MetaEdit) of the gut microbiome in vivo & directly modifying unculturable immune-modulatory SFB bug in the small intestine. 🦠🧬🛠️ https://t.co/6NYx3lQpMq
science.org
Although metagenomic sequencing has revealed a rich microbial biodiversity in the mammalian gut, methods to genetically alter specific species in the microbiome are highly limited. Here, we introduce...
3
11
54
📷📷📷New paper! (with @OpenAI) 📷📷📷 We trained weight-sparse models (transformers with almost all of their weights set to zero) on code: we found that their circuits become naturally interpretable! Our models seem to learn extremely simple, disentangled, internal mechanisms!
18
34
395
We are actively recruiting for two positions at the interface between biology and generative design. Backgrounds of particular interest are in protein biochemistry/evolution and synthetic genomics/biology. Please consider joining us! 1/n
8
92
348