Aditi Merchant
@aditimerch
Followers
771
Following
120
Media
9
Statuses
31
BioE PhD student @stanford in the Hie Lab // ML for SynBio
Joined April 2020
What if we could autocomplete DNA based on function? Today in @Nature, we share semantic design—a strategy for function-guided design with genomic language models that leverages genomic context to create de novo genes with desired functions.🧵 https://t.co/P5qVJB3qIY
16
154
590
Nature research paper: Semantic design of functional de novo genes from a genomic language model https://t.co/vB53a2eIGM
nature.com
Nature - By learning a semantics of gene function based on genomic context, the genomic language model Evo autocompletes DNA prompts to generate novel genes encoding protein and RNA molecules with...
2
14
45
Generative AI meets the genome
arstechnica.com
Genes with related functions cluster together, and the AI uses that.
0
4
7
Semantic design, our method leveraging contextual relationships between genes for function-guided biological sequence design, is now out in @Nature! This work was led fearlessly by @aditimerch, who inspires all of us in the lab every day. She carried out this immense project
What if we could autocomplete DNA based on function? Today in @Nature, we share semantic design—a strategy for function-guided design with genomic language models that leverages genomic context to create de novo genes with desired functions.🧵 https://t.co/P5qVJB3qIY
0
8
37
Published today in @Nature, @aditimerch & researchers from the @BrianHie lab report that the large-scale genomic model, Evo, is capable of using surrounding genomic context to produce novel, functional genes, enabling an emergent approach they've termed 'semantic design'.
4
25
91
Today in @Nature, in work led by @aditimerch, we report the ability to prompt Evo to generate functional de novo genes. You shall know a gene by the company it keeps! 1/n
7
103
543
Check out this amazing work by the incredible @aditimerch and team!! Prompting DNA language models a la guilt-by-association lets you design things that function with low seq id
What if we could autocomplete DNA based on function? Today in @Nature, we share semantic design—a strategy for function-guided design with genomic language models that leverages genomic context to create de novo genes with desired functions.🧵 https://t.co/P5qVJB3qIY
0
2
12
Context can steer Evo to generate multi-gene interactions (toxins and anti-toxins, anti-CRISPRs) that function in the lab. Many of the functional sequences have very low sequence similarity to any natural gene. Read the thread and paper Congratulations @aditimerch!
What if we could autocomplete DNA based on function? Today in @Nature, we share semantic design—a strategy for function-guided design with genomic language models that leverages genomic context to create de novo genes with desired functions.🧵 https://t.co/P5qVJB3qIY
0
11
85
This was all possible because of the support of my incredible PI @BrianHie and my amazing labmates @samuelhking and @exnx. I’m forever grateful to be surrounded by people who inspire me to be a better scientist. To learn more, check out the paper:
nature.com
Nature - By learning a semantics of gene function based on genomic context, the genomic language model Evo autocompletes DNA prompts to generate novel genes encoding protein and RNA molecules with...
1
2
23
Together, this work suggests that genomic sequence models can meaningfully generalize beyond characterized natural evolution. Looking forward, we hope that semantic design can serve as a starting point for function-guided design and optimization of genes across biology.
2
1
17
Beyond providing novel sequences for functions of interest, SynGenome can be used to predict the roles of domains of unknown function, reveal functional associations across prokaryotic biology, and catalog chimeric proteins with unique domain combinations generated by Evo.
2
1
15
Semantic design achieved high experimental success rates (up to 50%) without structural conditioning or fine-tuning. To explore semantic design more broadly, we created SynGenome, a database of generations from millions of prompts. https://t.co/bALWJyROqG
evodesign.org
100 billion base pairs of AI-generated genomic sequence
1
2
18
Next, we designed anti-CRISPR (Acr) proteins. Evo generated functional Acr proteins that protected against spCas9, despite some having no sequence or predicted structural similarity to known Acrs. This further supported the idea Evo could generalize based on context alone.
1
1
11
We next asked if semantic design could co-design more evolutionarily diverse sequences. Focusing on toxin–antitoxin systems, we successfully generated a functional RNA antitoxin, a de novo toxic gene, and broadly neutralizing antitoxins. Many had <30% sequence identity to nature.
1
1
13
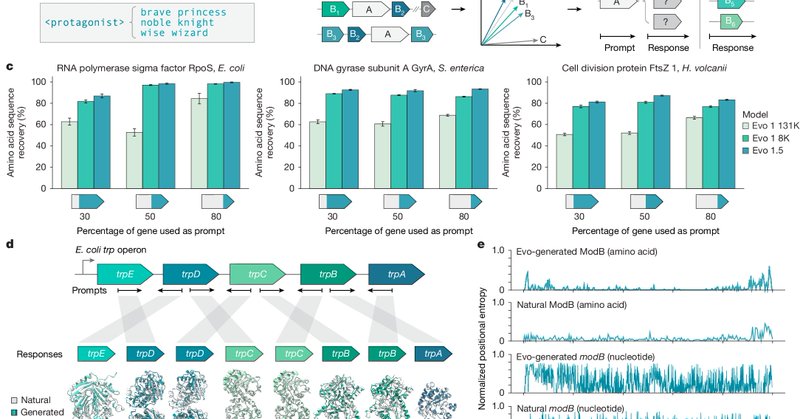
We first tested if Evo understands genomic context. Given partial sequences of conserved genes, we show that Evo can achieve near-perfect amino acid sequence recovery and complete entire operons bidirectionally, all while still producing diverse underlying DNA sequences.
1
1
18
Genomic language models like Evo can leverage this: by prompting with natural genomic context containing genes related to a function of interest, we can ‘autocomplete’ sequences with novel, diverse generations enriched for similar functions. We call this semantic design.
1
1
15
Just as word meaning emerges from context—"you shall know a word by the company it keeps"—prokaryotic gene function is tied to genomic context. This guilt by association, where related genes cluster in operons, has led to the discovery of molecular tools like CRISPR, BGCs, etc.
1
2
20
In recent years, we’ve seen immense progress in leveraging generative AI to accelerate biological design. However, using these models to produce diverse sequences with desired high-level functions remains challenging.
1
2
24
De novo antibody design with experimental success rates that require testing only tens of candidates! Such an inspiring success from the incredibly hardworking Germinal team— huge congrats!
The ability to design antibodies against any protein of interest has major implications for medicine, biotech, and basic science. Today, we introduce Germinal, a pipeline for epitope-targeted de novo antibody design achieving 4–22% success rates with efficient experimental
1
2
49
Evo-designed genomes are here!! HUGE congrats to @samuelhking for fearlessly bringing this project to life. Check out the thread to learn more! ⬇️
Many of the most complex and useful functions in biology emerge at the scale of whole genomes. Today, we share our preprint “Generative design of novel bacteriophages with genome language models”, where we validate the first, functional AI-generated genomes 🧵
2
7
36