David R. Liu
@davidrliu
Followers
76K
Following
10K
Media
678
Statuses
3K
Professor at Harvard and the Broad Institute. Our lab integrates chemistry & evolution to illuminate biology & enable new therapeutics. IG/Threads: @davidrliu1
Massachusetts, USA
Joined November 2008
Today in @Nature we report a new prime editing strategy that can rescue a common cause of many genetic diseases in a disease-agnostic manner. This approach converts a redundant endogenous tRNA into an optimized suppressor tRNA, enabling a single prime edit to rescue premature
33
381
1K
The Technion will award the prestigious Harvey Prize to two leading American scientists, Prof. David R.Liu and Prof. Chad Mirkin, for groundbreaking advances in nanotechnology and medicine. Their pioneering work is already transforming genetic therapies, diagnostics, and
4
9
42
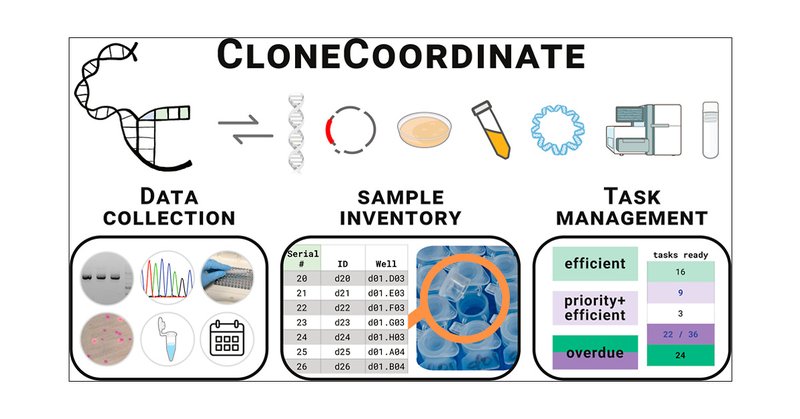
Delighted to report that our group's passion project over the last 6+ years to make DNA cloning more accessible, efficient, and scalable using a software-assisted workflow called CloneCoordinate is now out in ACS Synbio! https://t.co/w8gtYdmCXG
pubs.acs.org
Custom DNA constructs have never been more common or important in the life sciences. Many researchers therefore devote substantial time and effort to molecular cloning, aided by abundant computer-a...
1
7
43
Excited for #Deaminet! Registration is open join leaders in RNA editing & deamination research,Secure your spot before we hit capacity: https://t.co/ciGX3U0qZ1 ( https://t.co/YSIqigg5JS). Top sponsors, amazing sessions, & a collaborative community. Don’t miss out bring your lab!
labs.uthscsa.edu
0
5
12
This work was co-led by @Sarah_E_Pierce and @stevenerwood, with key contributions from @KeyedeOye, @meirui_an, @NicholasKrasnow, @emilyzhang326, @AdityaRaguram, Davis Seelig and Mark Osborn. Congratulations to the team! (15/15) https://t.co/LaRi2AwAeM
nature.com
Nature - A new strategy that uses prime editing to convert an endogenous tRNA into a suppressor tRNA shows therapeutic potential for multiple genetic diseases that are caused by premature stop codons.
1
5
47
President Trump is right. To stay ahead in the global AI race, America needs a unified federal framework. One that promotes innovation without overbearing regulation. A smart, cohesive national policy will ensure that American ingenuity leads the world in the age of A.I.
49
60
501
We hope PERT will provide a foundation for treating many diseases caused by premature stop codons with one of a small set of prime editing agents. More broadly, we hope this approach will catalyze additional allele-agnostic and disease-agnostic strategies that eventually help
0
2
9
To evaluate PERT in vivo, we prime edited the orthologous tRNA gene in mice to install an optimized mouse suppressor tRNA. In a mouse model of Hurler syndrome, this single edit restored IDUA activity to therapeutic levels and rescued all measured disease phenotypes to normal or
2
3
35
Using this optimized prime editing system, we rescued premature pathogenic premature stop codons in six human cell models of Batten disease, Tay–Sachs disease, and Niemann–Pick disease type C. The same prime edit, installing a single engineered suppressor tRNA, restored
1
3
32
Perhaps for this reason, together with the multiple mechanisms cells evolved to prevent readthrough of natural termination codons, we did not observe readthrough at any of the >4,000 protein-coding genes naturally terminated with a TAG stop codon in human cells upon PERT
1
5
29
Everyone deserves a world without impaired driving. Learn how Waymo is working to make our roads safer.
0
0
9
Importantly, prime editing the optimized suppressor tRNA into a single genomic tRNA locus resulted in underexpression, rather than overexpression, of the suppressor tRNA. Indeed, the suppressor tRNA was generated at levels ~2.5-fold lower than that of the endogenous Leu-TAA-1-1
1
2
20
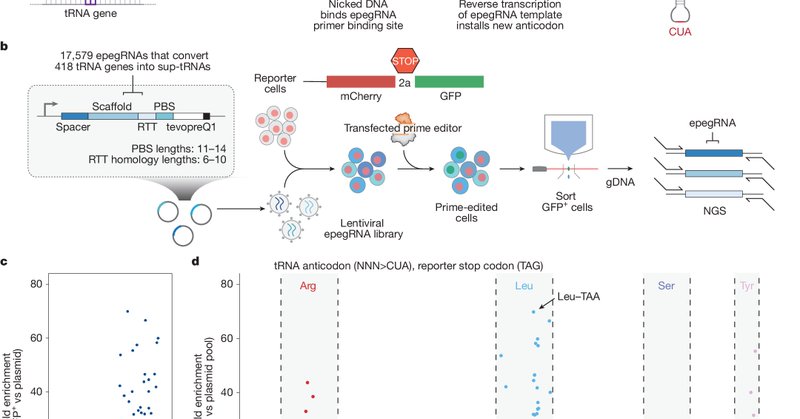
To optimize installation of these engineered suppressor tRNAs, we performed self-targeting lentiviral screens covering nearly 200,000 editing conditions, surveying diverse epegRNA architectures and prime editor variants. Editing outcomes closely mirrored endogenous editing,
1
1
21
The best-performing anticodon-only suppressor, tRNA-Leu-TAA-1-1, achieved ~7.5% readthrough of TAG (the most common premature stop codon) in the reporter assay. While encouraging, higher levels of suppression may be needed to rescue many premature stop codons. To discover more
1
4
25
To identify suitable endogenous tRNAs for PERT, we performed a high-throughput prime editing screen of all 418 human tRNA genes, rewriting each anticodon to complement each of the three stop codons and quantifying readthrough with a GFP reporter. A subset of endogenous tRNAs
1
6
23
A growing number of young Americans are being pushed by social media and doctors to to take hormones and undergo surgery—now they're detransitioning and sounding the alarm.
166
1K
3K
Here we report prime editing-mediated readthrough of premature termination codons (PERT). PERT uses prime editing to convert a dispensable endogenous tRNA gene into a highly optimized suppressor tRNA, enabling permanent readthrough of premature stop codons from a one-time
2
8
38
Importantly, many of the 418 high-confidence human tRNAs genes are functionally redundant, and humans can tolerate substantial variation in tRNA gene sequence (including anticodon mutations!) or copy number. Some tRNA families are missing entirely in healthy individuals. This
1
5
31
Suppressor tRNAs have long been studied as potential treatments for diseases caused by premature stop codons. But their lifetime dosing poses a major challenge, and their typical need to be present at high levels to achieve efficient suppression can perturb global translation.
1
6
32
24% (>52,000) of pathogenic mutations in the ClinVar database are premature stop codons, which are a cause of ~30% of known genetic diseases. A single suppressor tRNA can read through premature stop codons and restore full-length protein across many genes. By acting on the shared
1
10
63
Base editing and prime editing can correct most pathogenic mutations. But despite collectively afflicting ~400,000,000 patients, the vast majority of the >200,000 disease-causing mutations are too rare to attract drug development interest, at least until dramatic regulatory
2
11
62
Base editing and prime editing are redefining what’s possible in genetic medicine. Pioneered by David Liu, these tools are now showing real patient impact, including a personalized therapy for baby KJ developed in under seven months. Read more: https://t.co/2G0f4cOX4Y
1
12
46
That @US_FDA is recognizing here the urgent need for regulatory strategies to evolve to match the pace of scientific advances in ways that streamline bringing the science safely to patients is incredibly important. I hope these new policies provide a foundation that will
Personalized therapies hold tremendous promise but challenge traditional models of drug and biologic development. The @US_FDA outlines a path to market entry for products where a randomized trial is not feasible. Learn more: https://t.co/kt9YyRP0Hg
8
59
291