Yongsub Kim
@Yongsub__Kim
Followers
3
Following
49
Media
1
Statuses
21
Associate professor @ University of Ulsan College of Medicine (UUCM), Asan Medical Center (AMC).
Songpa-gu, Republic of Korea
Joined December 2022
Happy to share our lab's preprint! 📄 We've developed base editors with SsdAtox, a novel DYW-like deaminase from Pseudomonas syringae, effective for both ssDNA and dsDNA! #GenomeEditing #CRISPR #BaseEditing @biorxivpreprint.
biorxiv.org
CRISPR-based cytosine base editors enable precise genome editing without inducing double-stranded DNA breaks, yet traditionally depend on a limited selection of deaminases from the APOBEC/AID or TadA...
0
0
0
We've previously reported PAM variable PEmax for correcting KRAS mutations, and now our constructs - PEmax-SpG, PEmax-SpG-P2A-hMLH1dn - are available on @Addgene! Check them out! #CRISPR #PrimeEditing .
0
1
2
RT @zhaoweiasu: 1. The title of the recently published paper, "Genotoxic Effects of Base and Prime Editing in Human Hematopoietic Stem Cell….
nature.com
Nature Biotechnology - Base and prime editors induce double-strand breaks, deletions and translocations in hematopoietic cells.
0
15
0
RT @NicoleGaudelli: First patient dosed with Beam-201! 💫
globenewswire.com
BEAM-201 Represents First Quadruplex-edited, Allogeneic CAR-T Cell Therapy Candidate in Clinical-stage Development and First Treatment with a Base Editing...
0
44
0
RT @davidrliu: Today we report in @CellCellPress a suite of engineered and PACE-evolved prime editor variants PE6a-g. These new prime edito….
0
193
0
RT @NatureBiotech: Strand-preferred base editing of organellar and nuclear genomes using CyDENT .
0
34
0
RT @NikoMcCarty: A typical human cell expresses ~150-400 transcription factors. These proteins bind to DNA to regulate gene expression. An….
0
47
0
RT @CommsBio: A #CRISPR prime editing approach from @Yongsub__Kim corrects all types of G12 and G13 oncogenic KRAS mutations using a univer….
0
1
0
RT @zhangf: Since the discovery of CRISPR-Cas systems, people have wondered if a similar type of system exists in eukaryotes. Today, we rep….
0
2K
0
RT @davidrliu: A new report in @NEJM from Waseem Qasim and coworkers describes the clinical outcomes of the first three T-cell leukemia pat….
0
42
0
RT @biorxivpreprint: Discovery of new deaminase functions by structure-based protein clustering #bioRxiv.
0
1
0
RT @sniprbiome: 🚨We have published a paper in @NatureBiotech!. Find out more about the development of a #CRISPR-therapeutic to eradicate E….
0
31
0
RT @davidrliu: Today we report in @NatureBiotech the development of dual-AAV in vivo delivery systems for prime editors that can support ef….
0
217
0
RT @NatureBiotech: A highly efficient transgene knock-in technology in clinically relevant cell types https://t.co/….
0
29
0
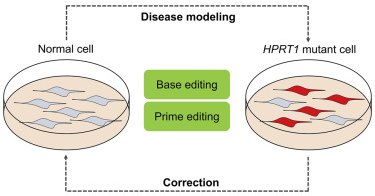
We report "Therapeutic gene correction for Lesch-Nyhan syndrome using CRISPR-mediated base and prime editing" in Molecular Therapy- Nucleic Acids. Congrat Gayoung & Ha Rim!.
cell.com
Lesch-Nyhan syndrome (LNS) is a rare genetic disorder caused by a deficiency of HPRT, and there are no fundamental treatment modalities for LNS. Jang et al. demonstrate that CRISPR-mediated base...
0
0
1
RT @edilytics: Congrats to See and @Yongsub__Kim on publishing their protocol on analyzing BRCA1 variants using Base Editors. Thanks for u….
link.springer.com
To date, methods such as fluorescent reporter assays, embryonic stem cell viability assays, and therapeutic drug-based sensitivity assays have been used to evaluate the function of the variants of...
0
1
0
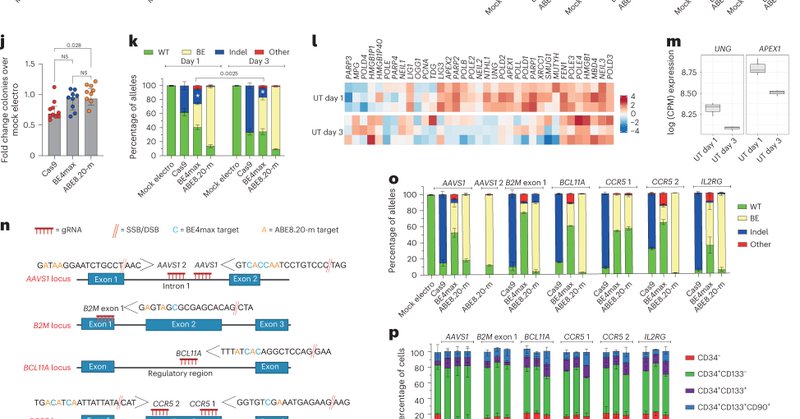
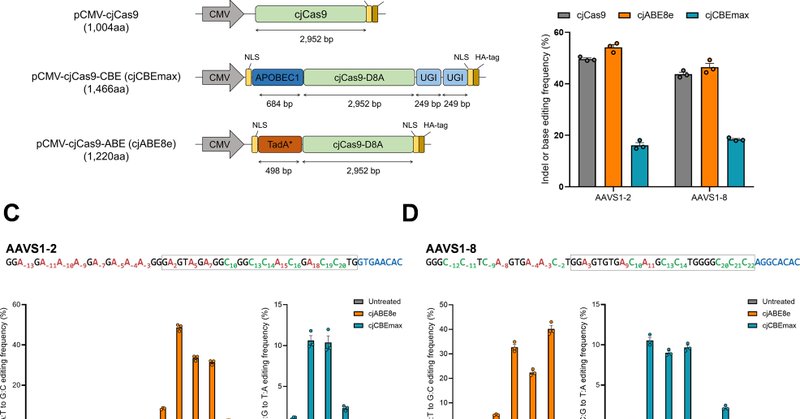
Today we report small size of CRISPR base editors using Campylobacter jejuni Cas9.
nature.com
Experimental & Molecular Medicine - Tiny genome editors that can fit inside a single delivery system could potentially improve the efficiency and specificity of the editing technology known as...
0
0
0
RT @biorxivpreprint: CRISPR prime editing for unconstrained correction of oncogenic KRAS variants #bioRxiv.
0
3
0