Edilytics
@edilytics
Followers
91
Following
13
Media
3
Statuses
252
Precision analytics for genome editing. Creators of CRISPResso2.
Joined February 2019
Thanks for using CRISPResso!.
Scientific sharing fuels progress. Our friends at @Edilytics developed CRISPResso, an open-source CRISPR analysis tool built for researchers. Together with OpenCRISPR-1, the future of customizable gene editing is more accessible than ever. Check it out:
0
0
0
We are thrilled to be a part of @BioUtah!.
🎉 Welcome, @Edilytics, to @BioUtah!. Their flagship product, CRISPResso, is the industry standard for analyzing genome editing events from NGS data, supporting diverse editing modalities with 1,650+ citations in academia & industry. 🔗 Learn more:
0
0
1
We are excited to attend @CSHL's CRISPR Frontiers conference next week. Let us know if you are going and we can talk all things genome editing analysis!.
0
0
0
Congratulations to @CRISPRTX and @VertexPharma for UK approval of Casgevy to treat sickle-cell disease and β-thalassaemia. Hope to continue to see more therapies brought closer to the clinic!.
nature.com
Nature - The landmark decision could transform the treatment of sickle-cell disease and β-thalassaemia — but the technology is expensive.
0
0
0
Happy #WorldCRISPRDay! To celebrate we would like to announce the launch of Join us in celebration by analyzing your #CRISPR experiment, happy analyzing!
0
0
1
Our team is excited to be at the @ESGCT congress next week! If you are going we would love to meet you in person, feel free to reach out so that we can connect.
0
0
0
Fascinating protocol from Changtian Pan @PCT71158462 & Yiping Qi constructing CRISPR-Combo systems for speed breeding of genome-edited plants. Thanks for using #CRISPResso in your analysis!.
nature.com
Nature Protocols - This CRISPR-Combo system enables efficient multiplexed orthogonal genome editing and transcriptional activation in plants. Here, the use of CRISPR-Combo is demonstrated for speed...
0
0
2
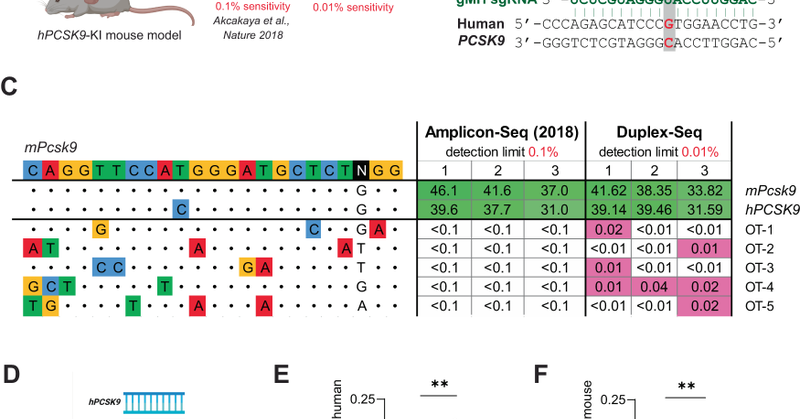
Congratulations to Bestas et al. on their paper illustrating a new nuclease with lower off-targets and translocations. Thanks for using CRISPResso!
nature.com
Nature Communications - SpCas9 unintended editing is a major concern. Here the authors report an off-target method using Duplex Sequencing with increased sensitivity for Cas9 mutation detection;...
0
0
1
Congratulations to Alex Reed, Benjamin Cravatt, @jfernandobazan, et al. on their groundbreaking paper utilizing #CRISPR technology to show the role of TMEM164 in ferroptosis. Thanks for using #crispresso2.
nature.com
Nature Chemical Biology - The function of a poorly characterized transmembrane protein, TMEM164, was annotated by integrated genetic dependency mapping, AlphaFold2 structural modeling and...
0
0
0
Congratulations to Mohr, Damas, Gudmand-Høyer, et al. on their groundbreaking @ACSSynBio paper utilizing #CRISPR technology for precise genetic engineering. Thank you for using #crispresso2! .
pubs.acs.org
CRISPR-Cas12a nucleases have expanded the toolbox for targeted genome engineering in a broad range of organisms. Here, using a high-throughput engineering approach, we explored the potential of a...
0
1
2
Congrats to Kaiqin She, Yi Liu, Qinyu Zhao, et al. on their recent paper on correcting mutations and phenotypes in retinal degenerative mice using split prime editing. Thanks for using #crispresso2.
nature.com
Signal Transduction and Targeted Therapy - Dual-AAV split prime editor corrects the mutation and phenotype in mice with inherited retinal degeneration
0
0
0
Congrats to Fuminori Tanihara, Maki Hiratat, et al. on their recent paper using electroporation and #CRISPR to cause an INS point mutation in pigs! . Thanks for using #crispresso2!.
frontiersin.org
Just one amino acid at the carboxy-terminus of the B chain distinguishes human insulin from porcine insulin. By introducing a precise point mutation into the...
0
0
0
Congrats to @kaisa_pakari @WittbrodtLab @_ThomThum_ et al. on their recent paper on de novo PAM generation to reach initially inaccessible target sites for base editing . Thanks for using #crispresso2.
journals.biologists.com
Summary: A combinatorial and sequential one-shot approach termed ‘inception’ is a twist on base editing that allows initially inaccessible nucleotides to be reached.
0
0
3
Congrats to Li, et al. on their recent paper developing prime editing. Thanks for using #crispresso2!.
nature.com
Nature Communications - Strategies to improve the specificity of nuclease-based prime editor (PEn) are needed. Here the authors report a 53BP1-inhibitory ubiquitin variant-assisted PEn platform...
0
0
0
Congrats to Jin, et al. on their recent paper on how to circumvent barriers to in vivo #CRISPR use. Thanks for using #crispresso2!.
genomebiology.biomedcentral.com
Background CRISPR-based toolkits have dramatically increased the ease of genome and epigenome editing. SpCas9 is the most widely used nuclease. However, the difficulty of delivering SpCas9 and...
0
0
0
Congrats to Liao, Chen, Hsiao, Jiang, et al. on their recent paper using #CRISPR base editing to potentially treat inherited monogenic blood disorders!. Thanks for using #crispresso2!.
nature.com
Nature Communications - Here, Liao and colleagues apply adenine base editor ABE8e and its PAM-less variant ABE8e-SpRY to β-thalassemia patient hematopoietic stem cells in the form of...
0
0
0
Congrats to @nicopmat #AhmedAllam et al. on their recent paper using high-throughput screen to analyze #primeediting outcomes. Thanks for using #crispresso2.
0
0
1
RT @PrimeMedicine: We're leading the next generation of gene editing and are excited to make our Twitter debut. Follow us to stay in the lo….
0
15
0