Toufic Mayassi
@TouficMayassi
Followers
249
Following
474
Media
15
Statuses
78
Mucosal immunologist drawn to the computational world | @banajabri lab graduate | Postdoc in @TheXavierLab at the @broadinstitute
Cambridge, MA
Joined June 2014
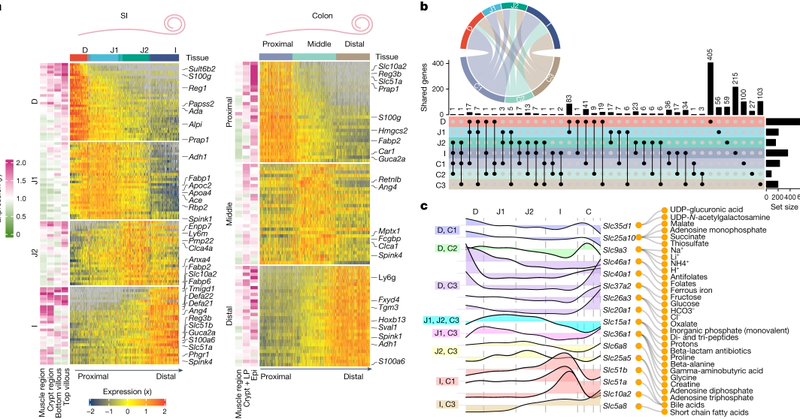
1/ Keen to share the story of my postdoc research in @TheXavierLab! We built a massive resource characterizing the spatial landscape of the gut, and yes, we dug deep into the biology too. If you’ve got a minute, let me walk you through the story. 🧵👇 https://t.co/sKL3yyCCKZ
nature.com
Nature - Intestinal regionalization is characterized by robust and resilient structural cell states and the intestine can adapt to environmental stress in a spatially controlled manner through...
21
70
295
Our most recent work on sphingolipids is now published in @cellhostmicrobe! https://t.co/9gr9ClbqUH Here we discovered a mechanism by which bacterial sphingolipids in outer membrane vesicles promote anti-inflammatory responses in immune cells, through activation of the mevalonate
3
34
128
Latest paper from lab, where Awad et al describe how cigarette smoke impacts that MAIT-MR1 axis and implications for lung diseases. In collaboration with Fairlie, Hansbro and Corbett labs https://t.co/hqlLiWHwsq
@MonashBDI @nhmrc @WaelAwadMoh @MonashUni @Jamie_Rossjohn @JExpMed
0
8
18
My book, An Intuitive Primer on Effective Functional Genomics Study Design, is published! I’d really appreciate it if you could help spread the word, and I’d love to hear your thoughts and feedback. It’s available on Amazon: https://t.co/VKR3l4RTSA
6
40
104
#WeekendRead! #LocationLocationLocation! @TouficMayassi @TheXavierLab &co show @Nature the regional specialization of the gut at the spatial (single cell) level: strong resilience after inflammation & ILC2 are key in middle colon to adapt to the microbiota
nature.com
Nature - Intestinal regionalization is characterized by robust and resilient structural cell states and the intestine can adapt to environmental stress in a spatially controlled manner through...
0
8
30
The intestine maintains a delicate balance in the body. This equilibrium is disrupted in parts of the organ in some diseases, but scientists don’t fully understand why. Now, researchers have created a blueprint of the gut that could address these questions https://t.co/d79fmBocTk
1
9
26
Huge congrats to Toufic and all! Been fortunate to witness this incredible piece of work come together over the years and all the hard work behind the scenes which led to so many surprising and interesting findings. Such as there is a very distinct middle colon region with
1/ Keen to share the story of my postdoc research in @TheXavierLab! We built a massive resource characterizing the spatial landscape of the gut, and yes, we dug deep into the biology too. If you’ve got a minute, let me walk you through the story. 🧵👇 https://t.co/sKL3yyCCKZ
0
2
14
1/3 Thrilled to share exciting new work in @Nature by @TouficMayassi, @li_chenhao, Åsa Segerstolpe, et al. Spatial profiling of mouse intestine reveals a landscape robust to perturbations in homeostatic drivers & adaptable in a spatially-restricted manner
nature.com
Nature - Intestinal regionalization is characterized by robust and resilient structural cell states and the intestine can adapt to environmental stress in a spatially controlled manner through...
0
30
105
It was such an experience working on this large-scale dataset and most importantly in the team with diverse backgrounds. I have learned so much from @TouficMayassi and other members of @TheXavierLab.
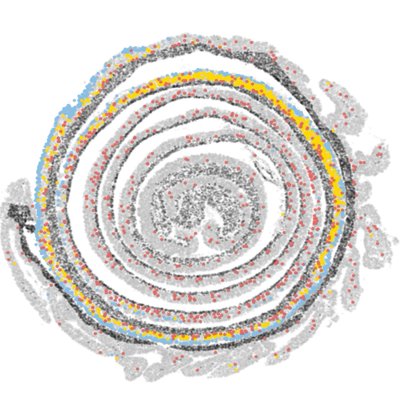
6/ Next challenge - reconstruct the tissue to its native state and unify segments within and across mice to facilitate downstream analysis and visualization. Enter the room @li_chenhao, a brilliant scientist that helped me supercharge the study along with my computational skills.
0
2
22
@li_chenhao @TouficMayassi @li_chenhao Hi, please find the unroll here: https://t.co/T2th6M7Fpd Talk to you soon. 🤖
threadreaderapp.com
@TouficMayassi: 1/ Keen to share the story of my postdoc research in @TheXavierLab! We built a massive resource characterizing the spatial landscape of the gut, and yes, we dug deep into the biology...
0
2
0
22/ Processed data and scripts can be found at https://t.co/SlQOakihII Data can also be viewed on the Single Cell Portal https://t.co/JQQqbJwi5x
https://t.co/ApgSaPlBqi
https://t.co/617Er6EP8a Read more about the story here:
broadinstitute.org
A spatial map of gene expression across the intestine reveals the organ’s remarkable adaptability.
2
0
7
21/ This work represents years of effort shaped by the guidance of mentors, support of colleagues, and encouragement from family and friends. Grateful to everyone who contributed, directly or indirectly. Excited to keep exploring the mysteries of the gut! Until next time...
1
0
4
20/ Our results demonstrate that intestinal regionalization is characterized by robust and resilient structural cell states and that the intestine can adapt to environmental stress in a spatially controlled manner via the crosstalk between immunity and structural cell homeostasis
1
0
5
19/ Remarkably, we turned off ANG4 expression in the middle colon. Using our Xenium panel, we showed the loss of the middle colon-adapted goblet cell population, demonstrating that ILC2s are essential for actively maintaining this adaptation.
1
0
3
18/ Intrigued by these results, we reached out to David Artis and his group @WeillCornell to test if ILC2s drive the middle colon’s microbiota adaptation. I drove down to NYC with my tools and together with the talented @HiroTheHero_PhD we turned the lights off on the mid colon.
1
0
4
17/ Together with my officemate of 4yrs and close friend @ericmichbrown we were then able to show ILC2 derived signals could induce the middle colon signature in ex vivo explants.
1
0
3
16/ We leveraged our sc data to compare all immune cell types across regions between SPF and GF mice and found only ILC2s showed a unique microbiota driven signature in the middle colon (B) which we could also validate with flow cytometry for a variety of markers.
1
0
2
15/ By now, you’re probably wondering what’s up with the middle colon? So were we. Mid colon genes like Ang4, Pla2g4c and Spta1 are linked to type II immunity tissue responses (helminth/protozoa colonization), yet we observed them in steady state and in absence of those drivers.
1
0
2
14/ Using our data to design a customized 480 gene panel we then leveraged high resolution Xenium technology to map all our cell states in space and validated the regional association of cell states as well as the middle colon adapted cells.
1
0
2
13/ We found unique structural cells defined each region (enterocytes, fibroblasts, and goblet cells) and strikingly the middle colon associated region (B) had unique subsets only found in the presence of the microbiota.
2
0
2