Xavier Lab
@TheXavierLab
Followers
3K
Following
266
Media
39
Statuses
516
Genes to functions. Microbiome in health & disease. Computational biology. Chemical biology. Innate & adaptive immunity. https://t.co/fsw4zVpmOE
Cambridge, MA
Joined September 2020
New in @NatureComms — collaborative work with the Knip & Orešič labs uncovers microbially conjugated bile acids in early life linked to immune development and type 1 diabetes risk. https://t.co/ImCa5y0ybl
@broadinstitute @MGHMolBio @MGBResearchNews
pubmed.ncbi.nlm.nih.gov
Emerging studies reveal that gut microbes can conjugate diverse amino acids to bile acids, known as microbially conjugated bile acids. However, their regulation and health effects remain unclear....
0
2
6
Register now for the International Human Microbiome Consortium (#IHMC) 2026, June 3–5 in Seoul, Korea! Join this global gathering of researchers for an exciting program highlighting the latest discoveries in human #microbiome science: https://t.co/lO0NbWRjGC
0
1
7
Registration now open for the International Congress of Mucosal Immunology (ICMI) 2026, Jul 13–17 in Montreal, Canada. https://t.co/QZ5WngJRK5 Looking forward to being part of this lineup of talks highlighting research at the interface of the immune system and mucosal surfaces!
socmucimm.org
ICMI provides an ideal opportunity to showcase your organization’s products and services and have direct interface with mucosal immunologists from around the globe.
0
2
5
Check out their findings from paired mother/infant microbiome & disease-specific metagenomic datasets! https://t.co/H9rehCP0l6 (2/2)
0
0
0
Solute carrier (SLC) transporters are implicated in many human diseases. In @jclinicalinvest, we show that efavirenz potentiates metal transporter SLC39A8 via a cryptic allosteric pocket—revealing new potential for therapeutic targeting. https://t.co/mWlqiNzGbs
0
1
3
Congrats to Jinjin Xu & co-authors – an image from their recent @JExpMed article that describes Ankrd55 as a modulator of T cell metabolism impacting Th17 responses was selected for the cover! Read the study here: https://t.co/sow57NlUzQ
@broadinstitute @MGBResearchNews
0
4
8
Great summary from the @MGBResearchNews team on our new Science paper about ENS–immune system interactions. Check it out:
0
2
4
Another one! Check out our new study in @ScienceMagazine! Using single-cell transcriptomics, we profile enteric nervous system responses to the microbiota and inflammation, identifying regulators of motor #neuron states and #gut transit time. https://t.co/DilPKBVYED
6
5
56
Out Now! Human immunodeficiency virus and antiretroviral therapies exert distinct influences across diverse gut microbiomes https://t.co/NF7raSAXqH
#HIV #GutMicrobiome #AntiretroviralTherapy
0
14
40
Congrats to all the authors! @broadinstitute
@MGBResearchNews
1
0
1
Excited to share new work in @NatureMicrobio! Using metagenomics, we show how #HIV infection and treatment shape the gut microbiome across geographic regions & find ART-induced phageome changes linked to inflammation and atherosclerosis. https://t.co/yEuvnuotl5
1
6
13
🔦Spotlight! The Korean Association of Immuologists International 2025 is convening Oct 30th-Nov 1st in Incheon, South Korea. KAI 2025 addresses current topics in immunology, encompassing both fundamental research & application to immunological diseases. https://t.co/8iOL8jaZIQ
0
0
0
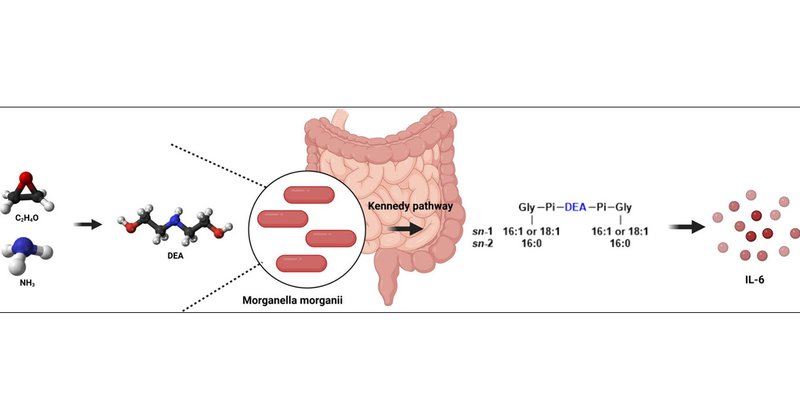
Our findings offer a new target that could be useful for diagnosing or treating certain cases of depressive disorder. They also provide a roadmap for probing how other members of the gut microbiome influence human health and behavior.
pubs.acs.org
A multifactorial association study detected a probable causal connection between the prevalence of Morganella morganii in the gut microbiome and the incidence of major depressive disorder (MDD) in...
0
0
1
In his “Harvard Medical School 2025 State of the School Address”, Dean George Daley highlighted our collaboration with the Clardy lab studying the connection between the gut microbiome and depression #Vimeo
2
1
2
0
0
3
The @GeneLayInst is excited to host its third #ReThinkNeuroimmunology meeting from October 14-16th, 2025. This year will focus on discussing major discoveries & breakthroughs in neuroimmunology, single cell genomics, immunology, & neuroscience. https://t.co/yrzuxIjHCT
1
4
21
via inefficient repression of a retinoic acid-mediated T cell gut-homing program (2/2) @broadinstitute
@MGHMolBio
@MGBResearchNews
0
0
3
Check it out now! Our latest effort to translate genetic risk variants to disease mechanisms revealed that the R448G missense mutation in nuclear receptor coregulator NRIP1 promotes tissue inflammation...(1/2) https://t.co/ckd7gA274E
pnas.org
Nuclear receptors (NRs) are crucial to integrate metabolite sensing and immune responses in the gut. NR-interacting protein 1 (NRIP1) is an importa...
1
2
4