Tim Coorens
@TimCoorens
Followers
1K
Following
491
Media
60
Statuses
392
Group Leader @emblebi | Former postdoc in @getz_lab @broadinstitute | Former PhD student @sangerinstitute | Lineage tracing, cancer genomics, human development
Joined November 2019
Very happy and excited to announce that I'll be starting my own research group at @emblebi! The group will focus on lineage tracing, somatic evolution and the origins of cancer. Interested in doing a postdoc in the group or know someone who is? Please reach out!
41
31
262
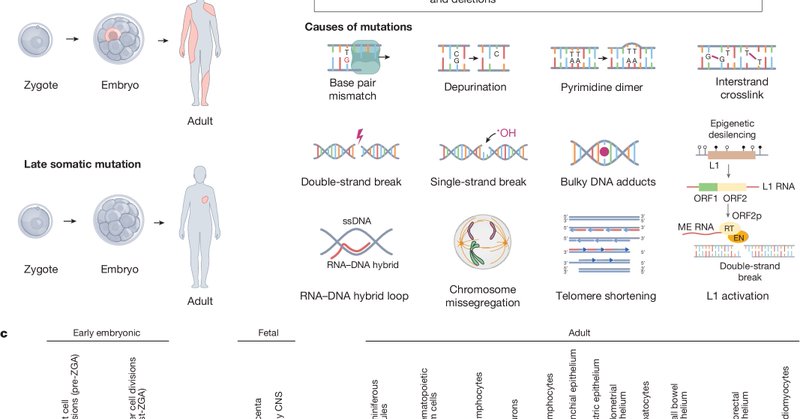
🧬 The SMaHT marker paper is now live in @Nature This landmark study characterizes somatic variation across 19 tissue types from 150 nondiseased donors, laying the groundwork for future discoveries in health, aging, and disease. Read the full paper: https://t.co/uXArDHEtHn
1
8
16
Last but not least, if you're interested in somatic mutations, new technologies and computational genomics - we're hiring a postdoc in my brand new lab @emblebi! Reach out if interested or you have any questions, and apply here:
0
0
4
This effort would be impossible without the work of many hundreds of people, including the scientists, the NIH staff, and - of course - the donors and their family. A very exciting time to study somatic evolution and I can't wait to see what the next years bring.
1
0
2
SMaHT will generate massive amounts of data, including short-read, long-read, duplex, single cell DNA, and donor specific assemblies, and much more. Check out our stellar data platform ( https://t.co/WYYmcUHibM), courtesy of @peter_j_park's lab - watch this space for future data!
1
0
3
Underpinning the efforts of the Network is a set of 150 donors from across the US and age range, collected by the NDRI, with many tissues samples for each donor - an absolutely unique collection and momentous effort. https://t.co/BJdvvCVU25
1
0
2
The SMaHT Network, funded by the @NIH, has over 300 members from over 50 different institutions all working together on developing new tools and technologies to detect somatic variation, analysing the data in the best way possible, and generating a reference catalog of mosaicism
1
0
2
The cells in our bodies constantly acquire mutations. But what are the patterns of mutations across tissues? How do mutations in normal cells lead to disease? These and other questions we will tackle within the SMaHT Network, now described in @Nature
https://t.co/5c1mfZOrOR
nature.com
Nature - The Somatic Mosaicism across Human Tissues Network aims to create a reference catalogue of somatic mosaicism across different tissues and cells within individuals.
4
41
131
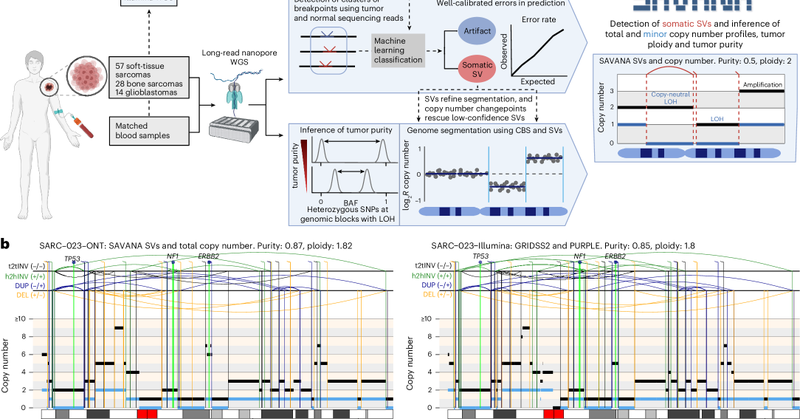
Thrilled to see #SAVANA out in @naturemethods🥳 SAVANA detects haplotype-resolved somatic SVs and copy number aberrations (SCNAs) and infers tumour purity & ploidy in clinical samples using long-read sequencing with or WITHOUT a matched germline control 👇 https://t.co/AdZhqHUsDb
nature.com
Nature Methods - SAVANA is a tool to detect somatic structural variants and copy number aberrations using long-read sequencing data, offering high sensitivity, specificity and compatibility with or...
3
28
134
1/ 🧵Check out our new study in @NatureGenet, which examines how precancerous conditions (MGUS/SMM) evolve into Multiple Myeloma, and offers new computational methods to predict progression risk and model cancer evolution. https://t.co/IHnN87QKTk
6
25
74
Call for Papers! @genomeresearch is now encouraging submissions for a Special Issue on The Genetics and Genomics of Somatic Mosaicism, Guest Edited by Drs. @giladevrony, @EAliceLee2, and @R_Rahbari. https://t.co/jKuoADbJec
#BOG25 #KSMosaicism25 #MITS25 #somaticmosaicism
0
16
38
Check out our new paper in @nature
https://t.co/cI3YCh7X1g. We report genetic mechanisms of neural tube defects in patients. Spina bifida is also known as meningomyelocele (MM). Prior family-based and association studies found few linked genes, so we took a different approach.
5
28
115
This new paper @Nature today on the genesis and evolution of stomach cancer goes against the alt-model https://t.co/hsjIMmVKBs
https://t.co/IABPCWKgK5
1
15
33
For the first time, scientists have analysed mutations in stomach lining tissue to reveal the earliest stages of cancer. The team also uncovered a potential new cause of stomach cancer, which needs further research 🔎 https://t.co/RZ5NXdMmHS
1
7
12
Happy to see our new paper out! A great collaboration with @jeshoag. We collected sequential sperm samples separated by decades, and used high-fidelity sequencing to measure germline mutation rates in individual men. https://t.co/jf9XkaKsZY
nature.com
Nature Communications - Mutations accumulate with age in the male germline, and can lead to genetic diseases in offspring. Here, the authors collect from individuals sequential sperm samples...
New paper from my group and @giladevrony’s! https://t.co/AGS3p55Qap Historically we measured new mutations in humans using trio studies (parents and offspring). This taught us that on average new mutations increase in offspring with paternal age. These trio studies showed
1
4
16
Thanks to all the donors and their families for making this study possible. And many thanks to Grace Collord, @KSaebParsy, Peter Campbell, @imartincorena, SY Leung, Mike Stratton and all other co-authors.
0
0
1
In short, the normal stomach shows a landscape of somatic mutations with many differences from those of other organs. Our findings provide insights into intrinsic and extrinsic influences on somatic evolution in the gastric epithelium in healthy, precancerous and malignant states
1
0
1
Because we used the LCM approach, we can map these driver clones to their location in tissues, which reveals expansions of mutant clones and local selective pressures. We find CTNNB1 mutant clones right next to one another, rather than evenly distributed.
1
0
0