Gilad Evrony
@giladevrony
Followers
708
Following
1K
Media
13
Statuses
289
🧬🧠 Center for Human Genetics & Genomics @nyulangone | @nyugrossman
New York, NY
Joined September 2013
Benchmarking of duplex sequencing approaches to reveal somatic mutation landscapes https://t.co/fjE6Ko34fe
#biorxiv_genomic
biorxiv.org
Detecting somatic mutations in normal tissues is challenging due to sequencing errors and the low allele fractions of post-zygotic variants. Duplex sequencing greatly reduces errors and can detect...
0
6
11
Our Center for Human Genetics & Genomics at NYU is recruiting new faculty investigators:
0
21
44
Please consider submitting to the special issue in Genome Research on somatic mosaicism!
Call for papers deadline is quickly approaching! More information on the topic of interests can be found here: https://t.co/1yL2FApzkk. Guest Edited by Drs. @giladevrony, @EAliceLee2, and @R_Rahbari. #ASHG25 #BOG25 #KSMosaicism25 #MITS25 #somaticmosaicism
2
2
8
Deadline extended to November 1!
Call for Papers! @genomeresearch is now encouraging submissions for a Special Issue on The Genetics and Genomics of Somatic Mosaicism, Guest Edited by Drs. @giladevrony, @EAliceLee2, and @R_Rahbari. https://t.co/jKuoADbJec
#BOG25 #KSMosaicism25 #MITS25 #somaticmosaicism
2
2
7
Highly accurate sequencing OR high throughput? We refuse to choose. Introducing paired plus-minus sequencing (ppmSeq), a method built to deliver both. Our latest paper with @UltimaGenomics 👀👀👀 See ⬇️ for a primer on the next frontier of genomics https://t.co/kl2KOaKziW
1
36
109
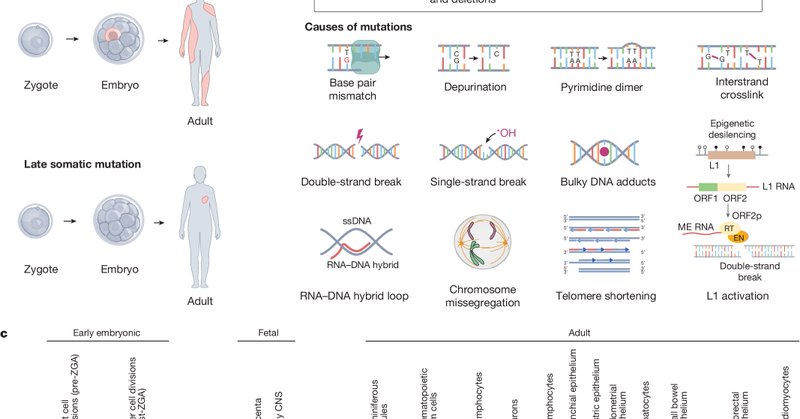
The cells in our bodies constantly acquire mutations. But what are the patterns of mutations across tissues? How do mutations in normal cells lead to disease? These and other questions we will tackle within the SMaHT Network, now described in @Nature
https://t.co/5c1mfZOrOR
nature.com
Nature - The Somatic Mosaicism across Human Tissues Network aims to create a reference catalogue of somatic mosaicism across different tissues and cells within individuals.
4
41
131
Call for Papers! @genomeresearch is now encouraging submissions for a Special Issue on The Genetics and Genomics of Somatic Mosaicism, Guest Edited by Drs. @giladevrony, @EAliceLee2, and @R_Rahbari. https://t.co/jKuoADbJec
#BOG25 #KSMosaicism25 #MITS25 #somaticmosaicism
0
16
37
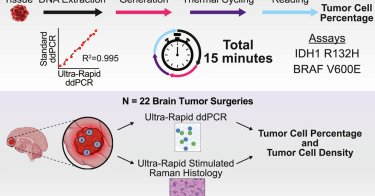
A new Ultra-Rapid droplet digital PCR developed by @giladevrony & @DanOrringerMD at @nyulangone may help surgeons detect cancer cells in real time. Early results suggest it could make brain tumor removal safer and more precise. Watch: https://t.co/yydYIzW4CP ✍️ @MedCellPress
2
5
21
Congrats to first author Ben Harrison (Rockefeller), and also to Emma Mizrahi-Powell (NYU). See also the concurrent paper by Kuehl, et al. on this new syndrome:
0
0
0
Just published--a new Fanconi anemia syndrome caused by variants in FAAP100 (FANCX)--a wonderful collaboration between our NYU Undiagnosed Diseases Program, the Smogorzewska lab (Rockefeller; @MaintainGenome) and Bruce Gelb and Cassie Mintz (Mount Sinai). https://t.co/UyMKh8Hytq
2
2
18
See also two awesome recent sperm sequencing studies from @R_Rahbari and @aaronquinlan. https://t.co/7w22KrnjYt
https://t.co/PaxWwB5uex
0
0
3
Kudos to @Amoolya1199, @CaitLoh, and @meihliu from my lab for their years of work on this, and huge thanks to the research participants, our collaborators, and our funders.
1
0
3
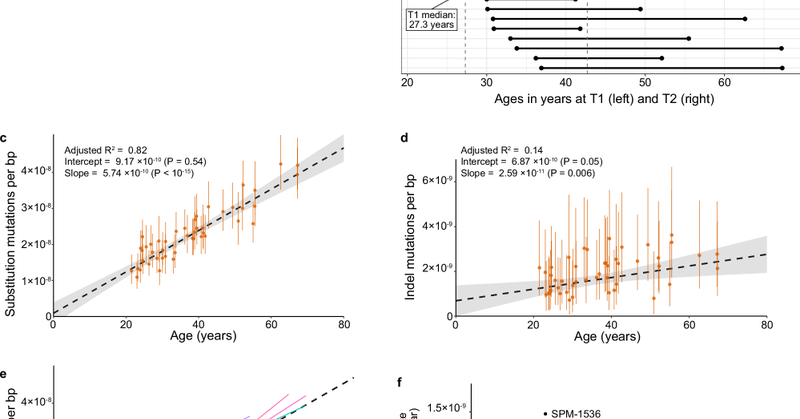
Happy to see our new paper out! A great collaboration with @jeshoag. We collected sequential sperm samples separated by decades, and used high-fidelity sequencing to measure germline mutation rates in individual men. https://t.co/jf9XkaKsZY
nature.com
Nature Communications - Mutations accumulate with age in the male germline, and can lead to genetic diseases in offspring. Here, the authors collect from individuals sequential sperm samples...
New paper from my group and @giladevrony’s! https://t.co/AGS3p55Qap Historically we measured new mutations in humans using trio studies (parents and offspring). This taught us that on average new mutations increase in offspring with paternal age. These trio studies showed
1
4
16
0
0
0
Our paper on Ultra-Rapid ddPCR (15 min tissue to result somatic mutation quantification) is now in @MedCellPress. A wonderful collaboration with @DanOrringerMD that succeeded because of the intrepid work of @ZacharyR_Murphy, Emilia Bianchini, and the team.
cell.com
Identifying a tumor’s molecular subtype during surgery and rapidly quantifying mutations in resected tissues intraoperatively could guide resection strategy and improve resections. Murphy, Bianchini,...
3
10
45
Writing the Fermi Paradox chapter of the book and man that topic just never gets old. Double the length of my 2014 post and updated with all the new ideas that have come out in the last decade. https://t.co/qq2G5IPYPk
waitbutwhy.com
The Fermi Paradox: There should be 100,000 intelligent alien civilizations in our galaxy — so why haven't we found any of them?
435
516
4K
Excited to see NYU Langone Health researcher @giladevrony's recent @Nature study highlighted in @NIHDirector's latest blog post! ⤵️
0
7
14
Thrilled to share the culmination of 4 years of #Fiberseq research enabling the accurate quantification of chromatin accessibility across the 6 Gbp diploid human genome with single-molecule and single-nucleotide precision. See thread for the main findings https://t.co/JEaTMohlxK
biorxiv.org
Most human cells contain two non-identical genomes, and differences in their regulation underlie human development and disease. We demonstrate that Fiber-seq Inferred Regulatory Elements (FIREs)...
3
22
73
We show some new findings since the pre-print, including APOBEC3A's ssDNA cytosine deamination pattern and detection of SBS1 precursor lesions. See our pre-print thread for more info: https://t.co/6OwqHNRaey Congrats to co-authors and thanks to our funders!
🧬 Preprint 🧬 Most mutations begin as a change in only one of the two DNA strands. To see these single-strand precursors of mutations, we developed single-molecule sequencing that achieves single-molecule and *single-strand* fidelity. https://t.co/8Eyznw7voq 🧵⬇️ 1/11
1
1
6