Pascal Sturmfels
@PascalSturmfels
Followers
408
Following
331
Media
9
Statuses
128
PhD student at @uwcse + @UWproteindesign working on machine learning for protein design, protein language models, etc.
Seattle, WA
Joined December 2019
Even though I probably spent more time in the lab playing chess than doing research, for some reason they gave me a PhD 🥳🥳
2
4
41
That's my designed protein on the cover of Science! Achievement unlocked!
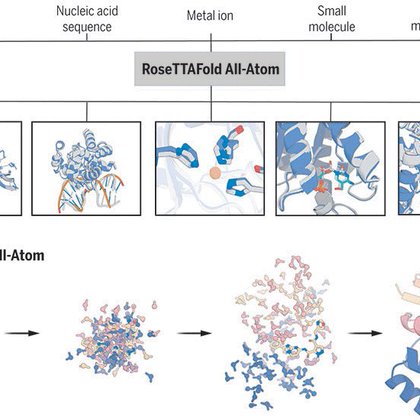
Researchers in Science present a next-generation protein structure prediction and design tool, #RoseTTAFold All-Atom, that can accept a wide range of ligands and covalent amino acid modifications. Learn more in this week's issue: https://t.co/PmmMUjxNaz
11
24
369
Researchers in Science present a next-generation protein structure prediction and design tool, #RoseTTAFold All-Atom, that can accept a wide range of ligands and covalent amino acid modifications. Learn more in this week's issue: https://t.co/PmmMUjxNaz
14
140
468
Specifying the input for cofolding models just got easier with my new gradio component "gradio_cofoldinginput". Try it here to generate inputs for RoseTTAfold2 All Atom predictions including covalent modifications:
huggingface.co
Our work on modeling and designing biomolecular assemblies is now in @ScienceMagazine. https://t.co/J0xnuTGGgZ RFAA Code: https://t.co/PkMLFH9iid RFdiffusionAA Code:
1
6
38
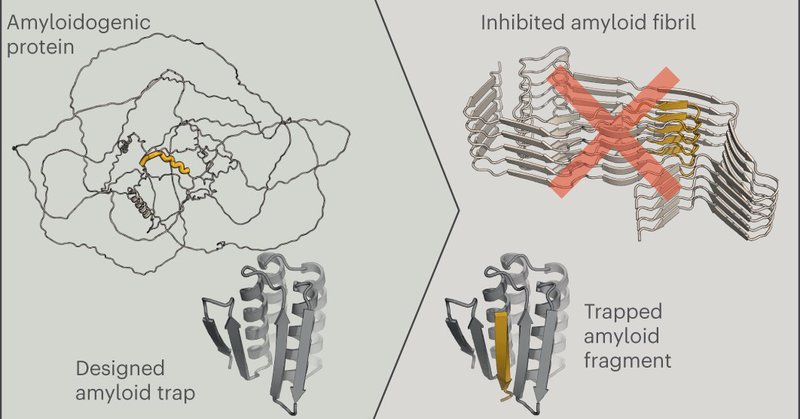
Super excited to share our paper “Design of amyloidogenic peptide traps” is out now! 🧠 https://t.co/Qmu9BpZiMA Special thanks to @eaandrzejewska , Danny Sahtoe and David Baker
nature.com
Nature Chemical Biology - An approach to design proteins that can capture amyloidogenic protein regions present in, for example, tau and Aβ42 has now been developed. These designer proteins...
2
26
131
We’re excited to share our preprint where we show, for the first time, the atomically accurate design of VHH antibodies!
16
154
616
🧵(1/7) Excited to share that our work, EquiformerV2, has been accepted to #ICLR2024. EquiformerV2 is the state-of-the-art on large-scale atomistic benchmarks -- OC20, OC22, AdsorbML, and ODAC23. Joint work with @bwood_m, @abhshkdz from @OpenCatalyst and @tesssmidt Paper:
2
32
164
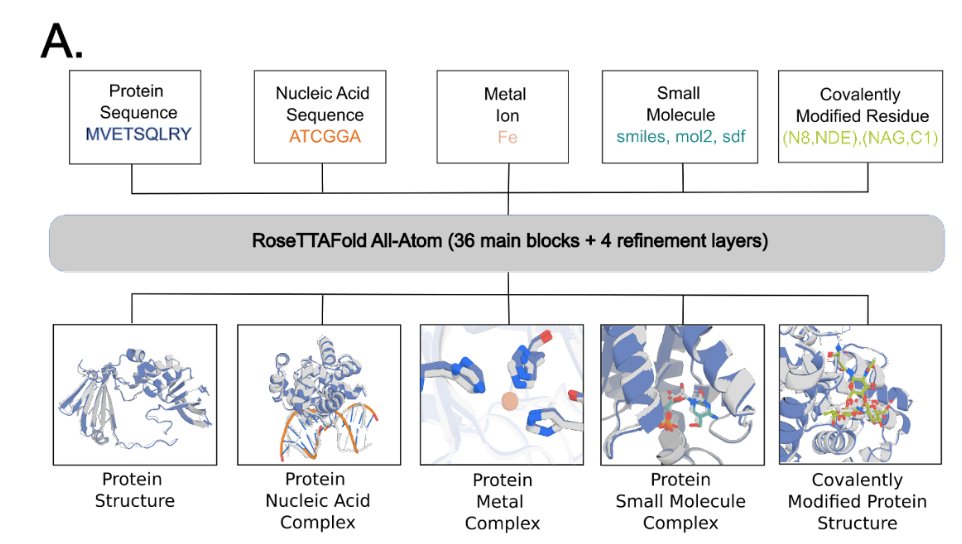
AlphaFold2 and RoseTTAFold are limited to predicting the 3D structure of proteins. A new all-atom method models complete biomolecular assemblies, including proteins, nucleic acids, small molecules, metals, and covalent modifications. #NBThighlight
science.org
Deep-learning methods have revolutionized protein structure prediction and design but are presently limited to protein-only systems. We describe RoseTTAFold All-Atom (RFAA), which combines a residu...
3
228
709
Our work on modeling and designing biomolecular assemblies is now in @ScienceMagazine. https://t.co/J0xnuTGGgZ RFAA Code: https://t.co/PkMLFH9iid RFdiffusionAA Code:
github.com
Public RFDiffusionAA repo. Contribute to baker-laboratory/rf_diffusion_all_atom development by creating an account on GitHub.
5
99
331
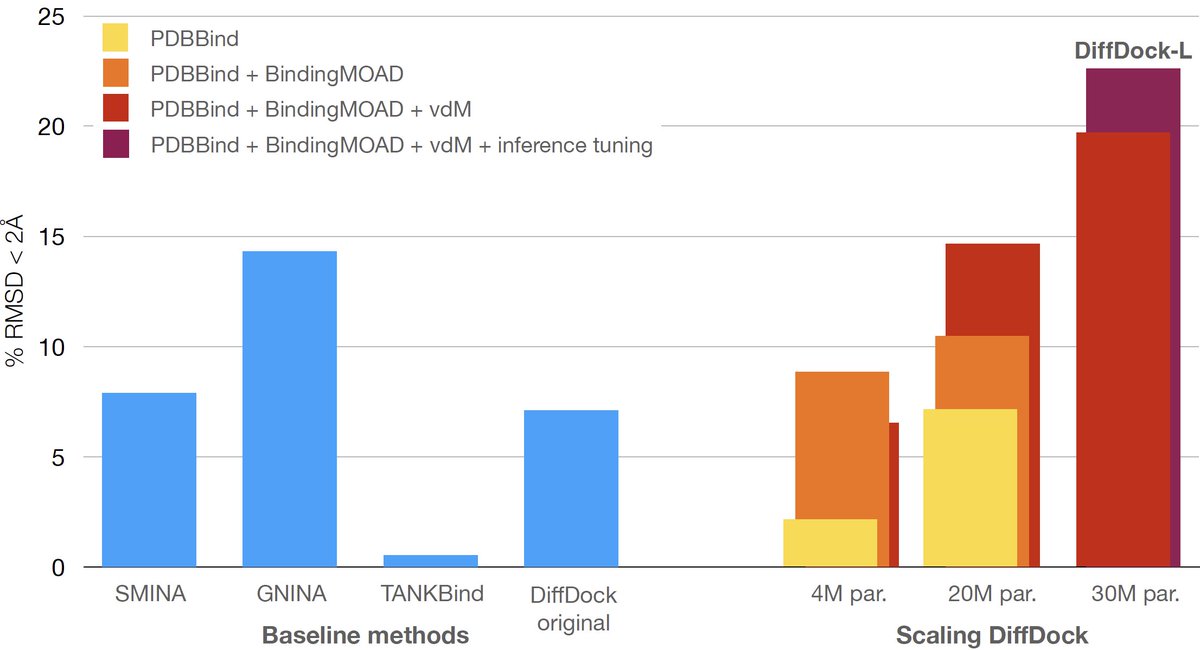
Excited to finally be able to share our ICLR work critically analyzing the capacity of deep learning docking methods to generalize and how to improve this (spoiler scaling, augmentation and RL)! With this, we release a new significantly improved version of DiffDock! A thread! 🧵
5
51
253
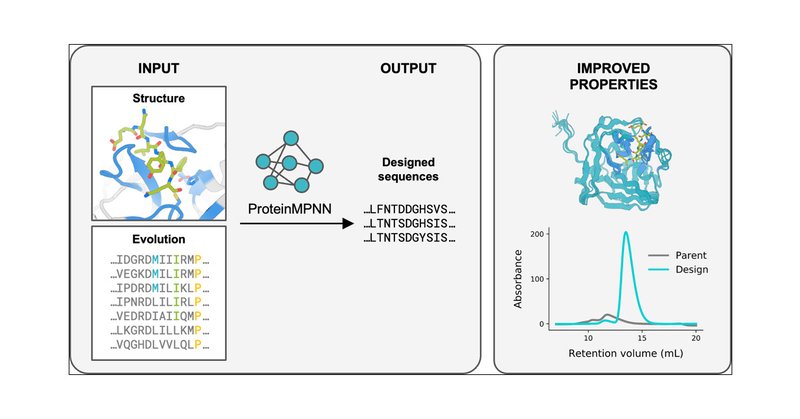
Our paper using ProteinMPNN redesign for improved properties is up on JACS! Huge thanks to Reyes, @ikalvet, Sam, and many others for their work. Shout out to @JustasDauparas for developing such a groundbreaking (and versatile!) tool. @UWproteindesign
https://t.co/bLm0jFuOvF
pubs.acs.org
Natural proteins are highly optimized for function but are often difficult to produce at a scale suitable for biotechnological applications due to poor expression in heterologous systems, limited...
3
73
261
looking for interns for two projects next summer, come work with me, @nc_frey and the rest of the @PrescientDesign team applying bleeding edge machine learning to fascinating biological data!
Come work with me, @samuel_stanton_ , and the fantastic @PrescientDesign @genentech team as a grad student intern this summer in NYC! https://t.co/TSOKpMP7bb
https://t.co/90Qyn2apoZ 🧵1/
0
9
33
Monday 11am Reading group: RosettaFold all-atom! https://t.co/Wh4UiZbbJE Let us discuss with @r_krishna3 what protein design niceties we can achieve with this. And also the AlphaFold all-atom "glimpse" :) Join on Zoom at 11am ET / 4pm UTC / 5pm CET: https://t.co/R8d1EHxLCx
1
22
135
Soo, this is a big one :) On Nov 20 we will have RosettaFold all-atom in the reading group with @r_krishna3! Quite amazing protein design capabilities. But that is my naive evaluation - join us and let's discuss and hear your insights! Sign up here: https://t.co/R8d1EHxLCx
1
34
168
Thrilled to announce the opening of the Srivatsan Lab ( https://t.co/mUPc2wHm0X) at the @fredhutch. The lab will be building new sequencing technologies to understand how our cells and bodies form over the course of development.
srivatsan-lab.com
Discovering, building, and communicating the wonders of biology to the world. We believe that our research can be a force for good in the world and that we have an obligation to conduct research...
The newest @FredHutch Basic Sciences lab opens its doors! The @SRsrivatsan Lab is developing new sequencing technologies to interrogate structure-function relationships during vertebrate development. The lab is growing and hiring at all levels:
45
95
612
RFdiffusionAA generating a small molecule binding protein against an experimental FXIa inhibitor (OQO), a ligand which is significantly different than any in its training dataset.
Very excited to share RoseTTAFold All-Atom and RFdiffusion All-Atom, methods for structure prediction and design of biomolecular assemblies! https://t.co/iG6rD0LHsi 1/n
1
57
230
RoseTTAFold updated to be All-Atom... biological assemblies containing proteins, nucleic acids, small molecules, metals, and covalent modifications ... and diffusion🤯 https://t.co/36EtN0IzoL
7
234
832
All-atom version of RoseTTAFold looks quite exciting!
biorxiv.org
Although AlphaFold2 (AF2) and RoseTTAFold (RF) have transformed structural biology by enabling high-accuracy protein structure modeling, they are unable to model covalent modifications or interacti...
3
24
120
We made RoseTTAFold that predicts everything in the PDB, not just proteins, the used it to make RFDiffusion that generates ligand-binding proteins, including a 10nM binder of digoxigenin. See @r_krishna3's thread for details.
Very excited to share RoseTTAFold All-Atom and RFdiffusion All-Atom, methods for structure prediction and design of biomolecular assemblies! https://t.co/iG6rD0LHsi 1/n
0
8
71
Very excited to share RoseTTAFold All-Atom and RFdiffusion All-Atom, methods for structure prediction and design of biomolecular assemblies! https://t.co/iG6rD0LHsi 1/n
10
167
577