Max Fottner
@MaxFottner
Followers
271
Following
3K
Media
4
Statuses
116
Postdoc in the @klanglab at ETH Zurich. Trying to shed light on the ubiquitin system using chemical biology.
München, Bayern
Joined February 2019
Happy that this is finally online! Genetic code expansion approaches to decipher the ubiquitin code @ACSChemRev Many congratulations to Vera, Max and Marco! @VeraWanka @MaxFottner @MarkoCigler 🥂🎉🥳 https://t.co/Y5oZ89bKgv
0
20
103
Thank you @GCEcenter @klang @AChemSynBio for the opportunity to share our work and hear what's happening with genetic code expansion at #GCE2024. And what a finish to the evening! 😎
1
1
7
Proud to see our 'detective story' on uncovering the MoA of Orpinolide published in its final form in @nchembio! Immensely grateful to @georg_e_winter, the Waldmann lab @mpimoph and @laraialab for being part of this fantastic collaboration!
2
9
34
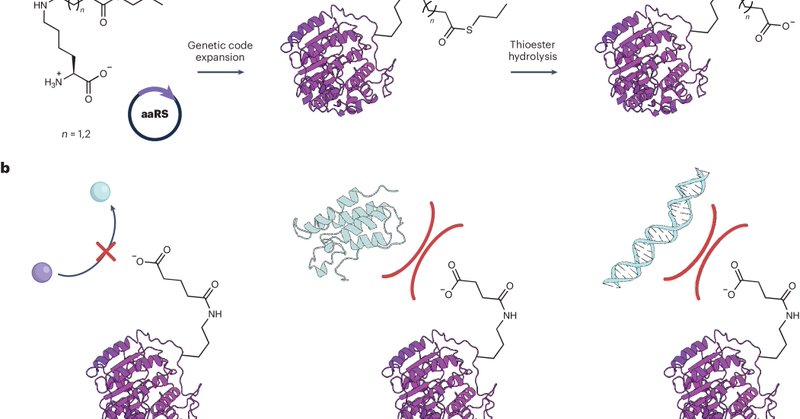
I'm happy to share our News & Views article featured in @NatureChemistry, highlighting the excellent work on site-specific incorporation of glutarylated and succinylated lysine residues into multidomain proteins via genetic code expansion by @klanglab! https://t.co/QzpQ2zaURy
nature.com
Nature Chemistry - Posttranslational modifications alter the structure and function of proteins. Now, genetic code expansion enables encoding of ε-N-succinyllysine and ε-N-glutaryllysine...
1
4
38
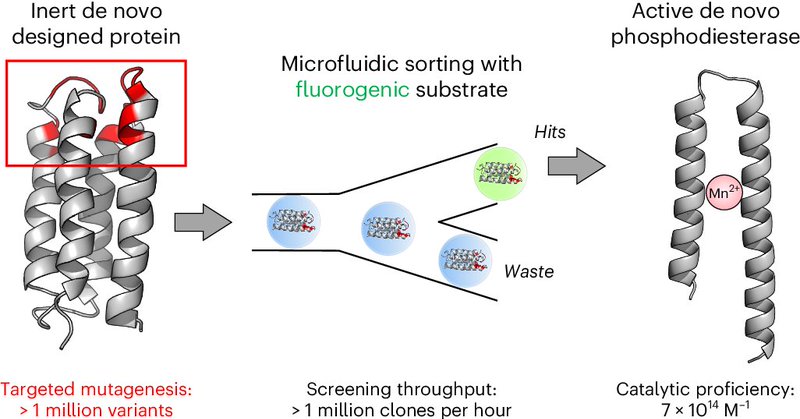
Check out our new paper @NatureChemistry where we use microfluidics to identify a mini enzyme from a de novo designed library giving a plausible scenario for emergence of enzymatic function at #OriginOfLife by #DirectedEvolution! https://t.co/6865tEhgVJ
nature.com
Nature Chemistry - Evolution separates complex modern enzymes from their hypothetical simpler early ancestors, which raises the question of how unevolved sequences can develop new functions. Here a...
1
18
113
Very excited to co-organize GCE2024 with @AChemSynBio and @Mehl_lab at @GCEcenter! Come to Corvallis, Oregon to hear about the newest in genetic code expansion! We have a stellar speaker lineup from academia and industry!👇 Retweet and register! https://t.co/g881i8cSks
gceconferences.org
Online GCE
🚨🚨All genetic code expansion enthusiasts, don’t miss the GCE 2024 conference this summer ( https://t.co/ItK2dA4Xi4)! Fantastic speakers both from academia and industry. Please retweet! @klanglab @Mehl_lab
0
6
28
Biocatalysts make chemical industry more sustainable but engineering enzymes is too slow! Check out our preprint @hollfelderlab where we show how microfluidics & AI can help: We rapidly sequence-function profile an IRED in microdroplets and use AI to accelerate its engineering⚡️
5
25
117
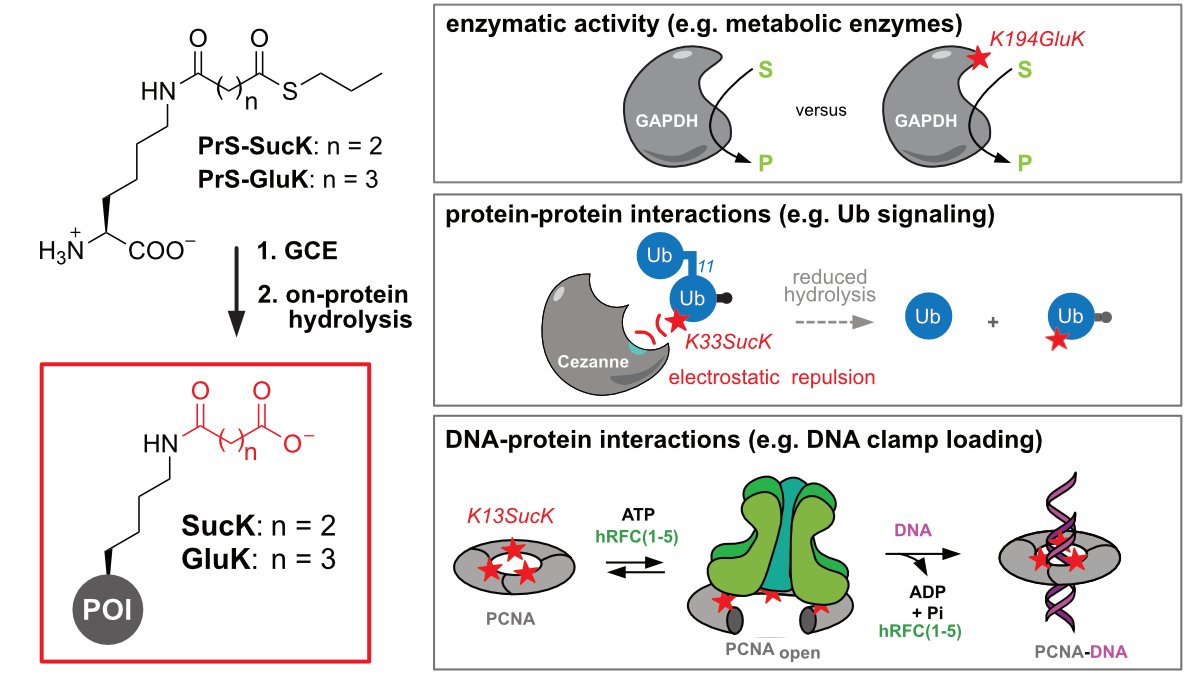
Genetically encoding negatively charged lysine PTMs succinylation/glutarylation and studying their funcional roles! Great team effort by first authors @ML_Jokisch and @Maria_Weyh! Congrats @Maria_Weyh @ML_Jokisch @MaxFottner and Anh! 🥳🤩🙌👨🔬🧑🔬 @klanglab
https://t.co/CKzQi1lavS
8
34
166
Check out our latest preprint @hollfelderlab presenting a versatile cascade for enzyme screening in microfluidic droplets! We hope coupled assay like this will make microfluidics more accessible reducing cost and speeding up sustainable biocatalyst discovery timelines 🌱⏰
Really excited to share the most recent preprint from the @hollfelderlab, a new fluorescence assay for NAD(P)H quantification that helps to bridge the gap between plate and droplet screening for enzymes with @MaxGantz_ and Jamie Klein. ( https://t.co/5ta0JYk813). When applying
1
1
17
We have a new doctor in the @klanglab! Well done to Kristina - Dr. Krauskopf - who successfully defended her thesis!🎓🥳👏 Many thanks to @SieberLab and Hubert Gasteiger for being on the committee at TUM! Very sorry for only being present via Zoom and missing the party fun 🤒🦠🙄
0
2
50
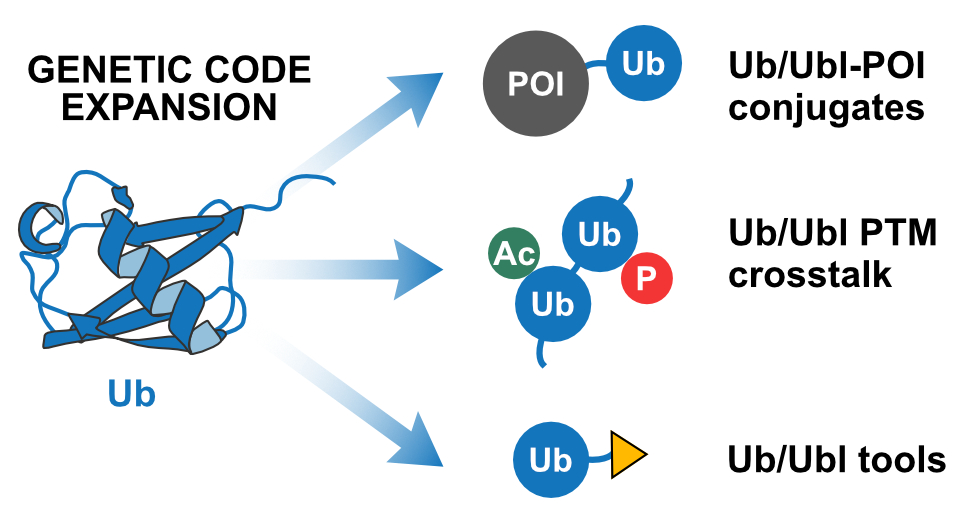
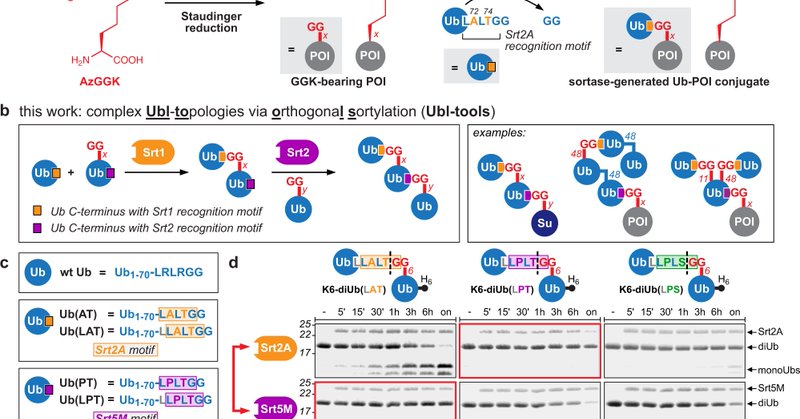
Amazing virtual talk by @klanglab at #ESBOC56 @ESBOC_Gregynog. She talked about her group's work on expanding the functions of proteins with non-canonical amino acids. Great applications in bioconjugations, crosslinking and defined ubiquitin conjugates.
nature.com
Nature Communications - Ubiquitin (Ub) and Ub-like modifiers (Ubls) can form chains of various topologies, but preparing defined chains for functional studies remains challenging. Here, the authors...
0
7
32
I am looking for a #Postdoc! Do you have #molecular_biology experience and are excited about developing top edge #drug_discovery modalities in a young team https://t.co/CxRthShr86? Join us @LACDR @LeidenScience Please RT! https://t.co/WfQIsFeXZc
pomplunlab.com
There is a labcoat waiting for YOU Bachelor & Master StudentsWe have several exciting M.Sc. projects available. The projects are in the field of early drug discovery and you will learn and appl…
0
8
10
@ERC_Research funded #postdoc position in my group We are looking for a molecular biologist excited to help developing synthetic tools that can control gene transcription. @UniLeiden @LACDR @LeidenScience Please RT!!! And apply :-) https://t.co/PknjqpvePp
0
6
3
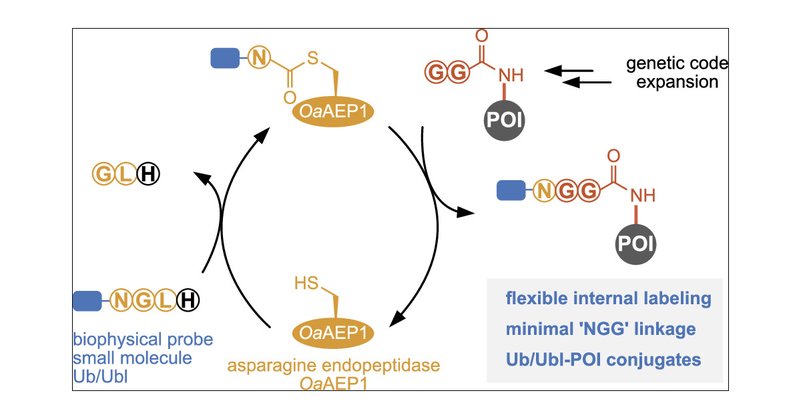
Check out our new paper for site-specific labeling proteins and building defined Ub/Ubl-protein conjugates using a plant Asp-endopeptidase. Great effort by @MaxFottner @jhmgrtnr @MaxGantz_ @RaMue1304 and @NkTimon! Congratulations to all!👩🔬👨🔬🤩🙌🥳
Site-Specific Protein Labeling & Generation of Defined Ubiquitin-Protein Conjugates Using an Asparaginyl Endopeptidase @ETH_DCHAB @ETH_en @klanglab @TU_Muenchen @MaxFottner #SiteSpecific #ProteinLabelling #Ubiquitin #Conjugates #Asparaginyl #Endopeptidase
5
25
82
Site-Specific Protein Labeling & Generation of Defined Ubiquitin-Protein Conjugates Using an Asparaginyl Endopeptidase @ETH_DCHAB @ETH_en @klanglab @TU_Muenchen @MaxFottner #SiteSpecific #ProteinLabelling #Ubiquitin #Conjugates #Asparaginyl #Endopeptidase
pubs.acs.org
Asparaginyl endopeptidases (AEPs) have recently been widely utilized for peptide and protein modification. Labeling is however restricted to protein termini, severely limiting flexibility and scope...
0
3
32
Sometimes we must create artificial things to understand nature. Kathrin Lang @klanglab, new professor of #ChemicalBiology #LOC, develops special tools and enables deep insights into complex biological systems. Learn more on June 1 #inaugurallecture & on https://t.co/TPZS7TWEkN
0
13
45
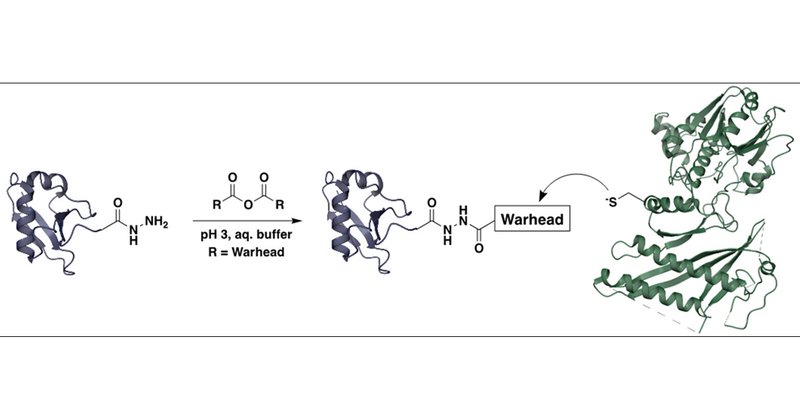
Just days after @StephenHacker2 ignited a debate on "warheads" in ABPs, @kattlm and @jakobfarnung bring you our collaboration with @jcornlab on a easy and powerful way to install them onto recombinant UFM1. https://t.co/Kbobg05UOD
pubs.acs.org
Aberrations in protein modification with ubiquitin-fold modifier (UFM1) are associated with a range of diseases, but the biological function and regulation of this post-translational modification,...
1
7
49
Small @klanglab reunion in Vienna after #ubiquitinfriends2022. Enjoying the best things beautiful Vienna has to offer with Vera, @MaxFottner and former PhD students @MarkoCigler and Anh, who are now both postdocs @CeMM_News. 🍸☀️🎠🐴🌇🍻
0
2
47
Such a pleasure to host this amazing crowd at our #ubfriends2022 #ubiquitinfriends2022 symposium!
1
14
76