Hollfelder Lab
@hollfelderlab
Followers
801
Following
112
Media
31
Statuses
122
Hollfelder research group @Cam_Biochem @Cambridge_Uni. Chemical and Cellular Synthetic Biology: From Mechanism to Droplet Microfluidics (and Back)
Cambridge
Joined October 2017
RT @iDEC2024: 🎉 The highly anticipated iDEC Festival 2024 is happening on October 26-27 in Cambridge! 📍 Three distinguished scholars will d….
0
5
0
Our take on the future of enzyme discovery: On synergy between ultrahigh throughput screening and machine learning in biocatalyst engineering - now published in Faraday Discussions
pubs.rsc.org
Protein design and directed evolution have separately contributed enormously to protein engineering. Without being mutually exclusive, the former relies on computation from first principles, while...
0
9
33
Thank you for continued wonderful collaboration(s)!.
@CamBiochem @hollfelderlab @Thav_Lab @MedCambridge @OPIGlets Great technology from @hollfelderlab for antibody discovery and, as always, pleasure to collaborate with them. Credit to Aleksei, Paul and Gwen from our side for protein work, biophysics and structural biology.
0
0
7
RT @CamBiochem: A Nature Biotechnology article by @hollfelderlab, @Thav_Lab, @HyvonenGroup, @MedCambridge, @OPIGlets establishes new techno….
0
5
0
RT @CamBiochem: A massive thank you to @francesarnold for visiting us last week and for delivering a wonderful Alkis Seraphim Lecture and t….
0
5
0
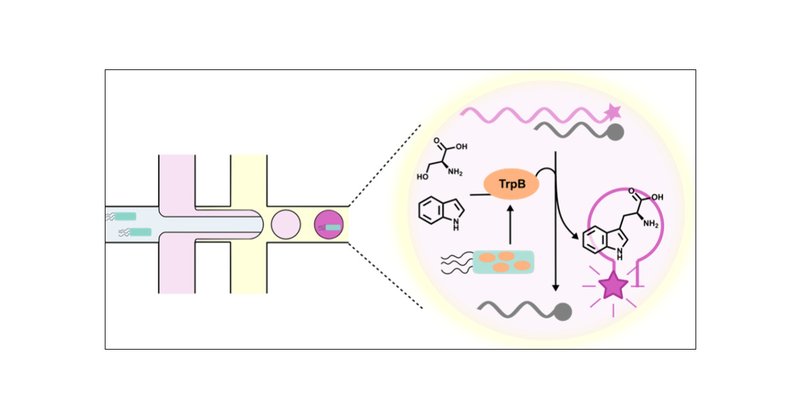
Remkes Scheele et al. make a new reaction accessible to large scale screening by tuning a DNA-aptamer to provide a fluorogenic read-out:.Ultrahigh Throughput Evolution of Tryptophan Synthase in Droplets via an Aptamer Sensor | ACS Catalysis
pubs.acs.org
Tryptophan synthase catalyzes the synthesis of a wide array of noncanonical amino acids and is an attractive target for directed evolution. Droplet microfluidics offers an ultrahigh throughput...
0
5
29
Wonderful breakfast chat on scientific life in general with @francesarnold, moderated by our researchers Lise Boursinhac, Josephin Holstein and Clara Munger. No doubt she has energy enough for a plenary talk this afternoon! .
0
2
16
RT @martanapio: Check out our preprint on YeastIT, a new system for continuous evolution, addressing the challenge of biased mutational spe….
0
10
0
SanntiS uses machine learning to identify biosynthetic gene clusters, achieves high precision and recall in genomic and metagenomic datasets, is extensively benchmarked and also experimentally verified: amazing new tool from @robdfinn @MGnifyDB @emblebi.
biorxiv.org
Natural products (bio)synthesised by microbes are an important component of the pharmacopeia with a vast array of biomedical applications, in addition to their key role in many ecological interacti...
0
0
2
RT @Bluetools_EU: Who are we?. Bluetools brings together five top European companies, eight university teams, and one private training orga….
bluetools-project.eu
A horizon Europe blue bioeconomy project making microbes research easier unlike traditional biodiscovery practices
0
1
0
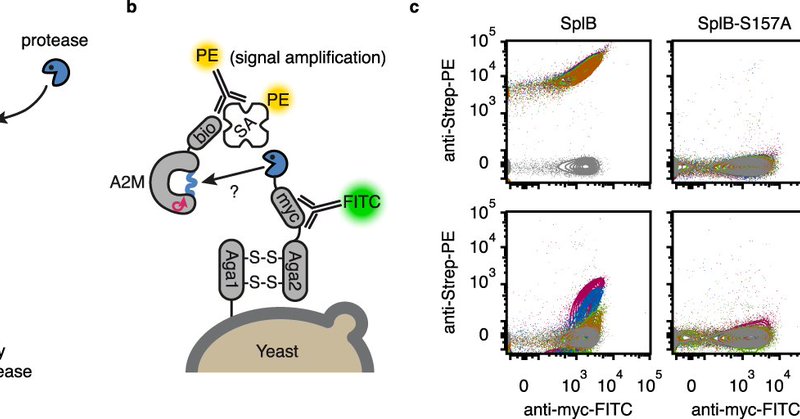
RT @LeDillingeur: Happy to see our paper from @hollfelderlab out in @NatureComms. In this work we describe a #DirectedEvolution method for….
nature.com
Nature Communications - Custom proteases find applications as therapeutics, in research and in biotechnological applications. Here, the authors establish a protease selection system based on...
0
8
0
Looking forward to interacting and collaborating with the BlueRemediomics initiative!.
ebi.ac.uk
Funding awarded for developing tools to harness marine microbiome data for biotechnological applications and ecosystem services.
0
0
3
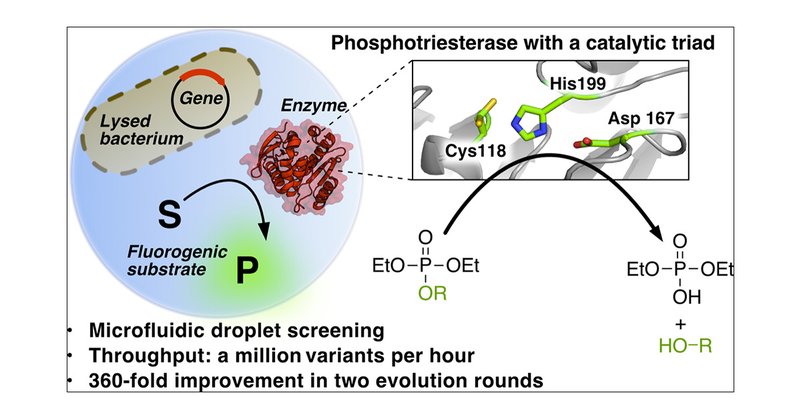
By evolving a promiscuous hydrolase recruited from a metagenomic library @JDSchnettlerF et al. show how a catalytic triad, more familiar from esterases and proteases, can also enable efficient phosphate triester hydrolyis.
pubs.acs.org
Finding new mechanistic solutions for biocatalytic challenges is key in the evolutionary adaptation of enzymes, as well as in devising new catalysts. The recent release of man-made substances into...
0
3
18
Stefanie Neun at al demonstrate (in collab with @HyvonenGroup) how droplet microfluidics can identify functions in metagenomic libraries that were not predicted by sequence-based methods. The quest for correct, comprehensive annotation starts here .
0
3
20
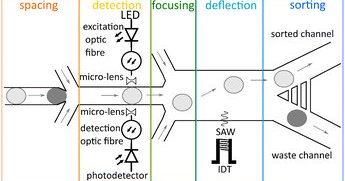
Esther Richter et al. integrate acoustic sorting with absorbance detection for more rapid ultrahigh throughput droplet screening. Great collaboration led by Thomas Franke’s group in Glasgow @EVOdrops_ITN .
pubs.rsc.org
Droplet microfluidics allows one to address the ever-increasing demand to screen large libraries of biological samples. Absorbance spectroscopy complements the golden standard of fluorescence...
0
2
12
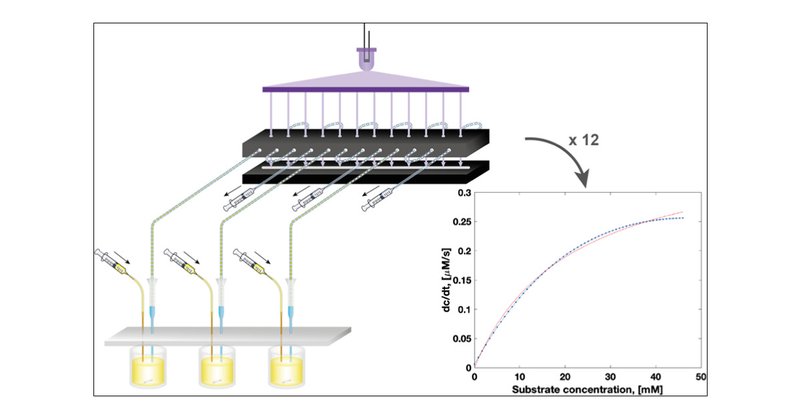
What to do when there are just too many mutants to be characterised - Stefanie Neun, @liisavanvliet & collab @FGieLab bring droplets to the rescue: HT Steady-State Enzyme Kinetics Measured in a Parallel Droplet Generation and Absorbance Detection Platform .
pubs.acs.org
Microfluidic water-in-oil emulsion droplets are becoming a mainstay of experimental biology, where they replace the classical test tube. In most applications, such as ultrahigh-throughput directed...
0
4
10