MatsNilssonLab
@MatsNilssonLab
Followers
1K
Following
181
Media
15
Statuses
131
Mats Nilsson lab at @Stockholm_Uni, part of @SciLifeLab, researching molecular diagnostics and in situ spatial transcriptomics 🔬🧬 🧫🔧
Joined August 2019
We're now also on Bluesky, sharing news from our lab. Follow us at the following handle! (.
nilssonlab.org
Our laboratory lies at the intersection between technical innovation and scientific exploration. We focus on the development, refinement, and application of...
0
0
0
RT @Dev_journal: Open-source, high-throughput targeted in situ transcriptomics for developmental and tissue biology. Read this Techniques &….
0
3
0
Many thanks to @CarolinaWahlby, Hanna Nyström and Peter Vermeulen for such a nice collaborative effort!🔎👩🔬.
0
0
0
More news!. 🚀 New research led by Maria Escrivà Conde & Axel Andersson from @CarolinaWahlby's lab , uncovers the cellular diversity and zonation patterns within the desmoplastic rim in #Colorectal #LiverMetastasis using #ISS 🔬💡 #CancerResearch .
1
3
8
🎉 Exciting news! Our latest work is now published in @Dev_journal! . The open-source solution for RNA-targeted in situ sequencing provides an accessible and efficient method for spatially resolved gene expression analysis and spatial multi-omics. 🔗
journals.biologists.com
Summary: We describe an improved open-source, flexible and customizable method and complete computational workflow for high-throughput multiplexed in situ transcriptomics, readily applicable to a...
0
8
15
🔎Interneuronal divesity in the human striatum uncovered!🧠 We are glad to contribute to the spatially-resolved characterization of striatum in an effort lead by @AMunozManchado and her team👩🔬 #spatialbiology #xenium.
Very glad to share our story on striatal human interneurons! Huge thanks for the great time that made it possible! @INiBICA_IIS @univcadiz @karolinskainst @NatureComms @SEBBM_es @SENC_.
0
3
6
New preprint alert! Work led by @PhyloBrain with contribution from our lab out in Biorxiv!.
NEW AVIAN BRAIN ALERT!.Check our research on the development of the pallium in birds🐣 and reptiles🦎. Comparative neurogenesis, scRNAseq and spatial-omics showing that the pallium of amniotes develops in different ways, but converge into similar circuits!
0
1
9
RT @martweig: Workshop alert: Together w @GioeleLaManno we organize on April 29 a 1day onsite event at EPFL about computational imaging bas….
0
22
0
And of course, none of this could be possible without the fantastic collaboration between the labs and the in situ sequencing facility @MatsNilssonLab & @GoCasteloBranco @karolinskainst @Stockholm_Uni @scilifelab .Congrats and great job to everyone involved!.N/N.
0
0
3
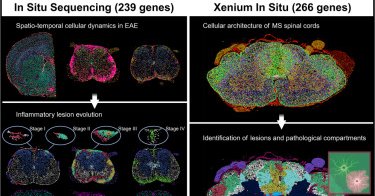
🚨 Our work on the cellular dynamics of lesion development in mouse experimental autoimmune encephalomyelitis (EAE) and human postmortem samples, now out in @CellCellPress!.Fantastic collaboration effort spearheaded by @chrislangseth and @PKukanja!. . 1/N.
cell.com
Spatial mapping of mouse and human multiple sclerosis neuropathology at a single-cell resolution unveils the cellular architecture and dynamics underlying disease evolution.
1
5
30
Glad to see technologies pioneered in the lab now accessible to the wider scientific community 🫡🤝.#spatialbiology #padlockprobes.
Dr. Mats Nilsson has spent his career developing new #spatialbiology and #singlecellspatial tools to break through technological limitations. See what he and Dr. Katarina Tiklova had to say about Xenium #InSitu and the past, present, and future of spatial!
0
4
19
🚨Job Alert🚨. Our Staff Scientist position at @sangerinstitute is now up and running. Do not hesitate. Apply today: LinkedIn:
0
3
11
🚨New preprint🚨.Who says spatially resolved transcriptomics solutions need to be extremely expensive 😤? We say, it does not have to be *that* complicated nor expensive. @lee_hower @chrislangseth @sergiomarco96 @Marcoooooooooo.
biorxiv.org
Multiplexed spatial profiling of mRNAs has recently gained traction as a tool to explore the cellular diversity and the architecture of tissues. We propose a sensitive, open-source, simple and...
This is our humble attempt to democratize in situ transcriptomics for the masses. Here in this preprint, we @MatsNilssonLab have put together an end to end solution for targeted in situ transcriptomics that is compatible with immunostaining and other labelling methods. 1/N.
0
9
34
🚨Looking to learn more about cutting edge spatial technologies?🚨. Join the #InSituSequencing Unit @scilifelab and @10xGenomics at Biomedicum on 11th May, 10am, introducing the new high performance Xenium spatial platform!. Registration link 👇.
scilifelab.se
Introducing high-performance subcellular spatial profiling made easy with Xenium In Situ. The Xenium platform reveals new insights into cellular structure and function by performing high-throughput...
0
5
18
Thrilled to share our latest collab publication in @natureneuro . Congrats to Xiaofei Li, @chrislangseth @lee_hower @dgyllborg and collaborators @adameykolab @slinnarsson @joalunatkthse Erik Sundström @HDCA_Sweden @scilifelab .
1
11
47
🔬 LAB MANAGER WANTED 🔬. The Nilsson Lab at @scilifelab & @Stockholms_univ is seeking a talented Lab Manager to join our diverse and international team! 💻🧪. Apply now:
0
10
9
In Situ Technology has come full circle! . Originally pioneered and developed in the Nilsson lab, in situ analysis / sequencing has come home in the form of two Xenium In Situ platforms from @10xGenomics. Xenium will be offered as a service by Spatial Omics platform @scilifelab
0
16
87
RT @sergiomarco96: Interested in the #Xenium In Situ platform from @10xGenomics? We've evaluated data quality, defined best practices and c….
0
35
0
Finally, congrats to @ASountoulidis and @sergiomarco96 for leading this @HDCA_Sweden effort and to the labs involved, including Samakovlis, @slinnarsson @joalunatkthse @Prof_Lundberg @CarolinaWahlby @jakmic2003 and Erik Sundström's lab.
0
0
5
These are some of our finding, but there is more biology in our data. For exploring our data, we put together a #Tissuumaps viewer with all the data. It's fast, interactive and easy to use. 9/N.
1
1
4