Mikaela Koutrouli
@MKoutrouli
Followers
461
Following
2K
Media
22
Statuses
327
PostDoc @genentech in the group of @MarioniLab | Interested in single-cells, deep learning, networks | Core member @scverse_team | https://t.co/wNWP4SbLVw
San Francisco, CA
Joined October 2018
Honoured to be part of this groundbreaking effort mapping the human DLPFC at single-cell resolution! Pushing the boundaries of neuroscience with incredible collaborators. Huge kudos to the #PsychAD team for making it happen!.
I’m thrilled to share one of the largest single-cell transcriptomic atlas of the human DLPFC with over 6.3M nuclei from 1,494 unique brain donors. Collaboration with @MKoutrouli Nick @g_e_hoffman @JaroslavBendl John @panos_roussos and #PsychAD Consortium.
0
0
17
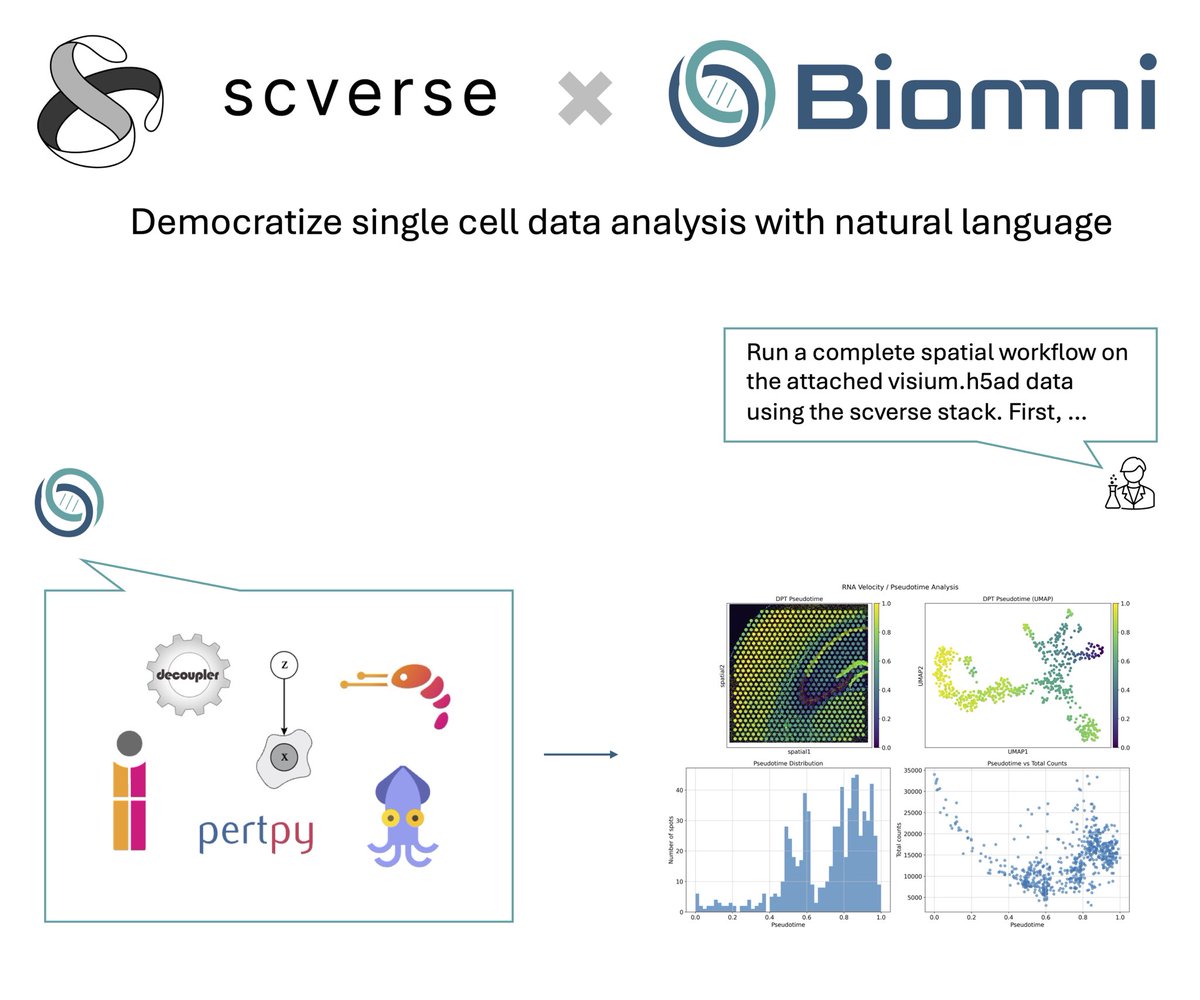
We're excited to collaborate with @ProjectBiomni to bring @scverse_team tools closer to a code-free future. This is an early-stage capability, and your feedback will shape what comes next. Try it out and help us build a more accessible analysis experience.
Thrilled to share @scverse_team x @ProjectBiomni! .Biomni agent now supports 10 scverse core packages including scanpy, squidpy, scirpy, pertpy, etc! .You can now use natural language to unlock complex single cell, spatial, and perturbation data analysis to generate novel
3
6
32
RT @saezlab: 🎉 The revised version of CORNETO, our unified Python framework for knowledge-driven network inference from omics data, is publ….
0
43
0
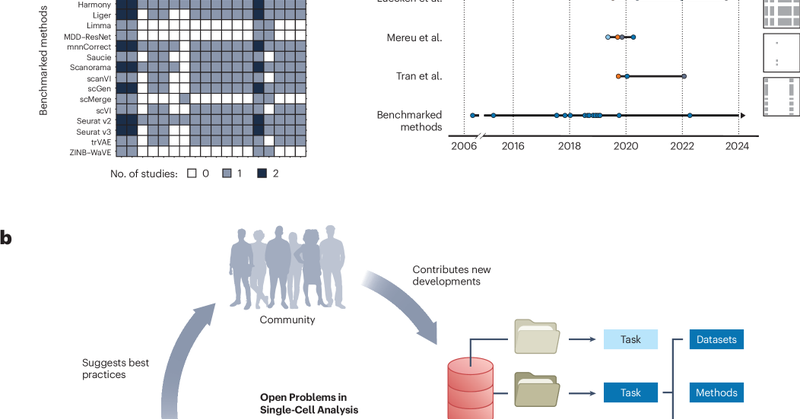
RT @fabian_theis: New OpenProblems paper out! 📝. Led by Malte Lücken with Smita Krishnaswamy, we present – a commun….
nature.com
Nature Biotechnology - Defining and benchmarking open problems in single-cell analysis
0
42
0
RT @saezlab: The final version of our multi-omics study on kidney fibrosis is out now (. Together w/ Pepperkok + Sa….
0
10
0
RT @NVIDIAHealth: 🆕🛠️ With the latest support for NVIDIA Blackwell GPUs and multi-GPU architectures, RAPIDS-singlecell, developed with @scv….
0
15
0
RT @anshulkundaje: @sara_mostafavi (@genentech ) & I (@Stanford) r excited to announce co-advised postdoc positions for candidates with dee….
0
47
0
RT @saezlab: 🔧 decoupler 2.0.6 is out thanks to @PauBadiaM, now as a core @scverse_team package This release inclu….
0
17
0
RT @hcwww_: Defining “what is good” is central to both AI and Science. Despite massive efforts towards building the Virtual Cell, our rec….
0
29
0
RT @labs_mann: Nano-scale, mega-impact: nanoPhos delivers 100× phosphoproteomics sensitivity, 70k sites from 1µg, 8k from 10ng! Powered by….
0
32
0
RT @jaanakprashar: Thank you so much @nalidoust for hosting the Tahoe Deep Dive hackathon!! Learned a lot and had so much fun. https://t.co….
0
4
0
RT @nalidoust: I am excited to announce the winners of our @tahoe_ai Tahoe Deep Dive hackathon with @huggingface. We created and released….
0
14
0
RT @honicky: Working with a large set of AnnData files on S3 is tough if you don't have the metadata indexed, so I created a tool to help.….
0
1
0
Honored to be part of the @scverse_team journey. Past, present, and future. All things single-cell and spatial. All open. Let’s keep going.
scverse turns 3!.What started as a shared vision for interoperable single-cell analysis has become a vibrant, global community. From AnnData to full multimodal pipelines, we’re building the future of everything single-cell and spatial omics, together. Here’s to what’s next!.
0
1
8
The @tahoe_ai hackathon just wrapped up! Such an inspiring environment with great energy, and genuinely fun people. Huge kudos to the @tahoe_ai team for organizing it so well. Extra proud to see the @scverse_team members in the teams that took all of the top 3 spots!.What’s next!
0
2
10
RT @FoltzPhd: Analyzing TCR or BCR single-cell data? Join the scverse community meeting this Tuesday!.
0
2
0