Liujuan Zheng
@LiujuanZ
Followers
27
Following
77
Media
0
Statuses
24
postdoc at Bange‘s lab @BangeBalcony at Uni Marburg and mpi-Marburg
Marburg
Joined April 2021
Fantastic work by the Narayan Lab connecting enzyme sequence space and substrate diversity!
CATNIP for the win! Read our newest work with the Gomes group- https://t.co/lILWdTxbVR
@Apatoneh @gabepgomes @daniil_boiko @AlisonNarayanUM
0
2
3
Specificities of chemosensory receptors in the human gut microbiota | PNAS great collaboration with @sourjik
pnas.org
The human gut is rich in metabolites and harbors a complex microbial community, yet surprisingly little is known about the spectrum of chemical sig...
0
4
7
It is out now 🥳🥂 I am very happy to be part of this nice collaboration in which we were able to characterize different bacterial chemoreceptors and their ligand specificities. Amazing teamwork with Wenhao Xu, @sourjik, @BangeBalcony, et. al 🤩 👇👇👇 https://t.co/JNRVTOO0DR
pnas.org
The human gut is rich in metabolites and harbors a complex microbial community, yet surprisingly little is known about the spectrum of chemical sig...
0
2
8
📢 A team led by Seigo Shima has revealed that #methane producing #microbes can reduce their need for #nickel when levels are low. The study sheds new light on the ecology of #methanogens and their impact on #climatechange. https://t.co/JHCauzQkCU
https://t.co/ahj10mZVjH
mpi-marburg.mpg.de
Methanogenic archaea have a significant impact on the global climate due to their role in producing almost all methane from natural sources. This well-known process relies on enzymes containing...
0
9
21
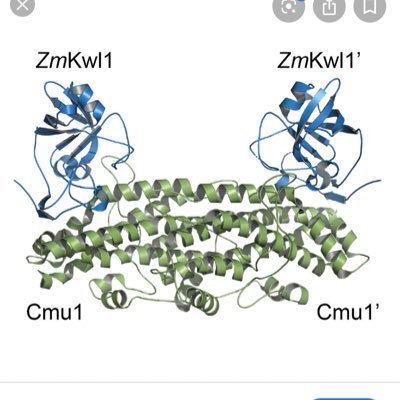
Molecular Microbiology | Microbiology Journal | Wiley Online Library
onlinelibrary.wiley.com
FlhG (orange, with activator helix in green) regulates peritrichous flagellation in Bacillus subtilis by interacting with the C-ring protein FliY (gray, N-terminus in red) and the cell wall regulat...
0
4
11
I'm truly excited to announce our new publication in @Nature unraveling a central picture of the Methyl-coenzyme M reductase (MCR) activation machinery and it's strong ATP dependency - kudos to @AnujR2d2 for the fantastic illustration! LINK: https://t.co/OmMRWWtNR8
8
39
118
I am more than happy to see this story to be out. We show another inhibition manner to Acetyl-CoA synthetase. Great thanks to mentor Gert and Joh. also great thanks to my best old friend Yifei as well as all contributed 🍀🍀🍀
A wonderful piece of work by Liu Zheng - enjoy the reading: Regulation of acetyl-CoA biosynthesis via an intertwined acetyl-CoA synthetase/acetyltransferase complex | Nature Communications @Uni_MR @mpi_marburg
2
6
10
I am so happy that the first story in Gert's lab is out now. We show how a fumarase family enzyme can catalyze decarboxylation of tran-aconitate to form the magic itaconate, Great thanks to my mentor Gert and Joh. And also my great master student Wei and all authors
Enjoy the read! Mechanistic and structural insights into the itaconate-producing trans-aconitate decarboxylase Tad1 | PNAS Nexus | Oxford Academic @Uni_MR @ERC_Research
0
1
6
Vier neue Vizepräsidentinnen und Vizepräsidenten gewählt - 2025 - Aktuelles - Philipps-Universität Marburg @Uni_MR
uni-marburg.de
Wiederwahl von Gert Bange und Sabine Pankuweit, Yvonne Zimmermann und Lara Zieß neu gewählt
1
3
14
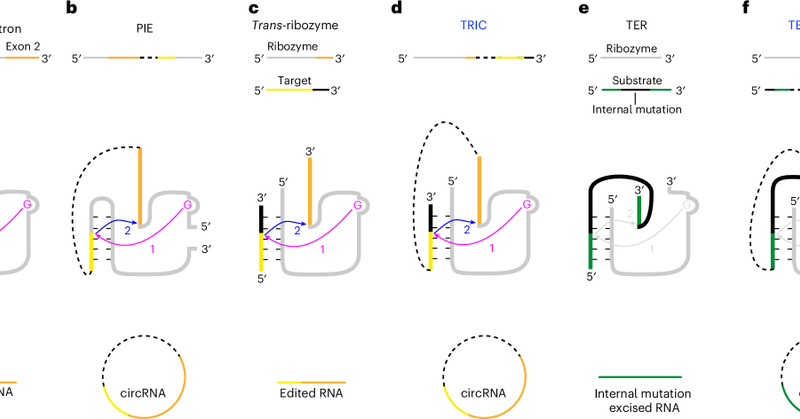
Excited to see this paper is out today: https://t.co/ATsLs2dsaf. We developed efficient circRNA synthesis methods and improved efficiency of Rolling Circle Translation by over 7000-fold. Huge thank to Venki and all colloboraters, especially @zuber_phil15 and @xueyan1104!
nature.com
Nature Biomedical Engineering - Trans-splicing-based methods that are independent of bacterial sequences and that allow the incorporation of RNA modifications enable the reliable synthesis of...
1
3
7
Novel enzyme starts with promiscuity. Excited to share our study that shows how #evolution recruits a single enzyme for multiple purposes, showcasing the plasticity and flexibility of #metabolism. Huge thanks to my colleagues at @erblabs & @mpi_marburg!
1
28
73
🚨Don't miss your chance to atttend the special one day symposium celebrating @NatProdReports 40th anniversary held @Helmholtz_HIPS! Join us in the celebrations with talks across natural products research 📅Register for free by October 18th▶ https://t.co/aYJqgaaNdK
0
7
8
Enjoy the reading: Mitochondria, Peroxisomes and Beyond—How Dual Targeting Regulates Organelle Tethering - great joint venture with my colleague, the twitterless Johannes Freitag @SYNMIKRO @Uni_MR
journals.sagepub.com
Eukaryotic cells feature distinct membrane-enclosed organelles such as mitochondria and peroxisomes, each playing vital roles in cellular function and organizat...
0
2
8
Polar confinement of a macromolecular machine by an SRP-type GTPase | Nature Communications great collaboration with @TheThormannDen @SYNMIKRO @Uni_MR
nature.com
Nature Communications - The SRP-type GTPase FlhF modulates the localization and quantity of flagella in bacteria through unclear mechanisms. Here, the authors show that FlhF interacts with the...
1
7
23
Alright everybody, what will undoubtedly forever be the lab's strangest discovery is now out in Nature. It's a protein that evolved to self-assemble into a Sierpinski fractal. https://t.co/zV5CDzbaGm 1/n
nature.com
Nature - Citrate synthase from the cyanobacterium Synechococcus elongatus is shown to self-assemble into Sierpiński triangles, a finding that opens up the possibility that other naturally...
15
197
885
Finally! After a long journey our latest NRPS engineering story is out at https://t.co/E77e2L7gn1 thanks to the hard work of several members from the @HelgeBode_lab
science.org
Bioinformatic analysis reveals sequence sites ideal for engineering in enzymes that produce non-ribosomal peptide natural products.
4
47
170
Oxepinamide F biosynthesis involves enzymatic D-aminoacyl epimerization, 3H-oxepin formation, and hydroxylation induced double bond migration.
pubmed.ncbi.nlm.nih.gov
Oxepinamides are derivatives of anthranilyl-containing tripeptides and share an oxepin ring and a fused pyrimidinone moiety. To the best of our knowledge, no studies have been reported on the...
0
1
4