Bode Lab
@HelgeBode_Lab
Followers
565
Following
23
Media
2
Statuses
35
Run by members of the Bode Lab. Doing all things related to natural products.
Marburg, Hesse
Joined March 2019
Happy to announce the first steps into NRPS/PKS engineering thanks to the work from Leonard and collaborations with the labs from Markus Kaiser and Michael Groll: Bioengineering of syrbactin megasynthetases for immunoproteasome inhib...
0
7
23
Leonard has applied the XUT NRPS engineering technology to solve the biosynthesis of the potent antibiotic odilorhabdin in collaboration with the Rolf Müller lab at @Helmholtz_HIPS. See his paper in @angew_chem:
onlinelibrary.wiley.com
The biosynthesis of the odilorhabdin biosynthetic gene cluster (BGC) was elucidated by engineering of the central non-ribosomal peptide synthetase (NRPS). Therefore, specific modules of the odilorh...
1
7
28
CRISPR now also available for Xenorhabdus and Photorhabdus for easy access to natural products and manipulation of their biosynthesis: Please see the paper from Alexander in Microbial Cell factories:
1
3
7
And here is another cool story about trans-AT PKS engineering in the same issue https://t.co/JdAOqTuDYK from the @piel_lab. Congratulations!!
science.org
Single-molecular tension sensors reveal cellular force loading rate during mechanotransduction.
0
6
40
Finally! After a long journey our latest NRPS engineering story is out at https://t.co/E77e2L7gn1 thanks to the hard work of several members from the @HelgeBode_lab
science.org
Bioinformatic analysis reveals sequence sites ideal for engineering in enzymes that produce non-ribosomal peptide natural products.
4
47
169
With our newest preprint on engineering NRPS we succeeded in optimizing our first-of-its-kind biocombinatorial technology. Read more here: Guidelines for Optimizing Type S Non-Ribosomal Peptide Synthetases
biorxiv.org
Bacterial biosynthetic assembly lines, such as non-ribosomal peptide synthetases (NRPS) and polyketide synthases, are often subject of synthetic biology – because they produce a variety of natural...
0
3
16
The latest update for NRPS engineering, based on a evolutionary breakpoint within thiolation domains is out:
biorxiv.org
Many clinically used drugs are derived from or inspired by bacterial natural products that often are biosynthesised via non-ribosomal peptide synthetases (NRPS), giant megasynthases that activate and...
0
7
33
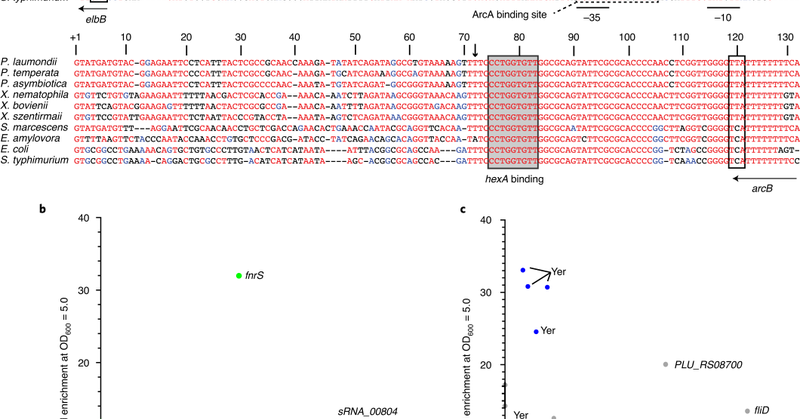
Next round in "the many things that regulatory RNAs can do" - this time: controlling natural-product synthesis. Fantastic collaboration with the Bode lab (@HelgeBode_Lab) published today in @NatureMicrobiol
nature.com
Nature Microbiology - A small RNA regulates the production of metabolites with roles in symbiosis and virulence in Photorhabdus and Xenorhabdus.
0
15
65
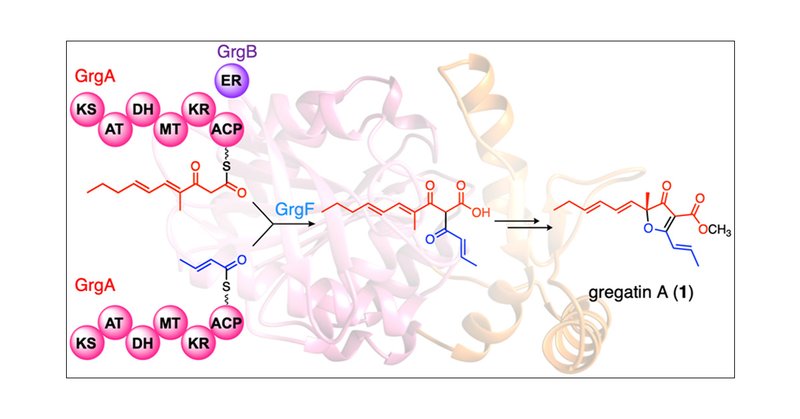
An awesome addition to polyketide biosynthesis. TE disguises itself as KS catalyzing branched polyketide formation. Read about Molecular Basis for the Biosynthesis of an Unusual Chain-Fused Polyketide Gregatin A
pubs.acs.org
Gregatin A (1) is a fungal polyketide featuring an alkylated furanone core, but the biosynthetic mechanism to furnish the intriguing molecular skeleton has yet to be elucidated. Herein, we have...
0
9
45
Luckily, despite severe jet lag, Helge’s given a clear, nice talk @SIMB_Microbio #NaturalProducts on NRPS engineering with a more efficient approach. Showing the advantages of artificial docking domains in splitting and reconstructing the megasynthetases
0
2
19
Finally... more than a module! Great NRPS work from the Schmeing lab: Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility
0
7
22
Who makes antibiotics and how do NRPS work? See the Grininger/Bode group performance for kids here:
0
0
8
🤔 could have submitted more
JOC invites you to take a look at this Note from first-time corresponding author Helge Bode & coworkers: (±)-Alternarlactones A and B, Two Antiparasitic Alternariol-like Dimers from the Fungus Alternaria alternata P1210 https://t.co/OTj8m9bxqz
0
0
8
We have new, hypermodified peptides for you! Check out our aeronamide (from Microvirgula) & polygeonamide (from deep-rock metagenome🤘🎸) story at @NatureChemistry
https://t.co/OwFWyBRhMu
#RiPPs
0
13
70
Natural products go RNA: The Natural Product Butylcycloheptyl Prodiginine Binds Pre-miR-21, Inhibits Dicer-Mediated Processing of Pre-miR-21, and Blocks Cellular Proliferation
0
0
11
Read Jörn‘s @nature #behindthepaper on how we developed the TransATor, a web tool for de novo structure predictions of trans-AT polyketides & why you need it
0
11
33
First time to achieve peptide ligation in water, offering a chemical perspective of chicken-and-egg problem for the origin of life
0
6
16
Nice work by the Stanley-Wall lab: Pulcherrimin formation controls growth arrest of the Bacillus subtilis biofilm
pnas.org
Biofilm formation by Bacillus subtilis is a communal process that culminates in the formation of architecturally complex multicellular communities....
0
0
1
Tom's amazing @NatRevChem review is finally out!! Learn more about hidden #enzymology, unprecedented chemistry & new approaches for exploring microbial dark matter at #naturalproducts
2
30
73