Jolien Perneel
@Jolien_Perneel
Followers
254
Following
971
Media
5
Statuses
145
Postdoctoral Researcher | Neurodegeneration, Lysosomes & FTD | Rosa Rademakers Lab, VIB-UAntwerp Center for Molecular Neurology
Antwerp - Belgium
Joined October 2013
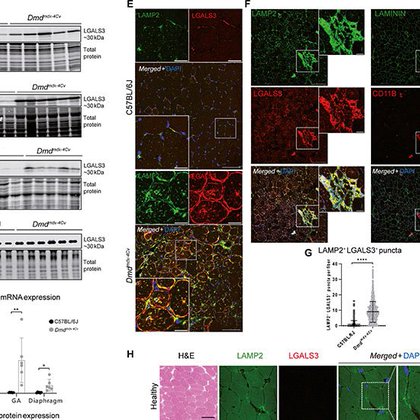
Lysosomal damage is a therapeutic target in Duchenne muscular dystrophy | Science Advances
science.org
Lysosomal damage contributes to DMD pathology and is a therapeutic target worth considering for current and future approaches.
0
11
39
And that’s a wrap - I successfully defended my PhD last week! Grateful for everyone who made this journey possible (- and meaningful). It feels a bit strange and surreal, yet incredibly rewarding to close such an important chapter of my life. Excited to see what comes next!
0
0
0
Almost there! 🎓 Excited (and a bit surreal) to be defending my PhD soon! Welcome to join the defense 👇
0
0
3
'Molecular hallmarks of excitatory and inhibitory neuronal resilience to #AlzheimersDisease' Isabel Castanho, Pourya Naderi Yeganeh...Rudolph E. Tanzi @RudyTanzi & Winston Hide @harvardmed @BIDMChealth
#CognitiveResilience #transcriptomics #genetics
https://t.co/xaaBvQyihh
0
10
21
The @progranulin_nav provides: 📍 Clinical FTD trial directory 🧬 Free genetic counseling + testing for eligible participants 📚 Resources for families + providers 🔗 https://t.co/KnZXV1f7Va
#FTDAwareness #FTDResearch #CureFTD #EndFTD #progranulin
0
2
3
Methylome analysis of #FTLD patients with #TDP43 pathology identifies #epigenetic signatures specific to pathological subtypes Cristina T. Vicente, Tejasvi Niranjan, Elise Coopman...Rosa Rademakers @CMN_VIB @MayoClinicNeuro
https://t.co/VojSighv76
0
8
15
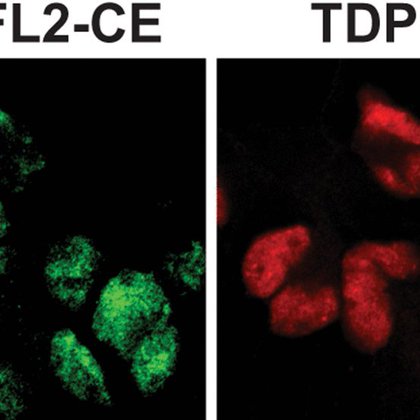
Seeded aggregation of TDP-43 induces its loss of function and reveals early pathological signatures https://t.co/2oKNWqYZdI
cell.com
TDP-43 pathology characterizes neurodegenerative diseases. Scialò et al. use in vitro-generated and patient-derived TDP-43 aggregates as seeds to show that seed-induced TDP-43 cytoplasmic aggregation...
0
6
22
Deciphering distinct genetic risk factors for FTLD-TDP pathological subtypes via whole-genome sequencing:
nature.com
Nature Communications - Here the authors identify TNIP1 as a risk factor for a fatal neurodegenerative disorder and discover specific genetic loci associated with the three main subtypes of this...
0
8
31
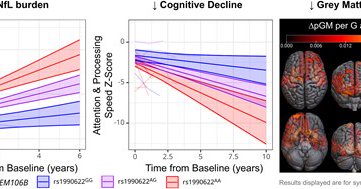
Very happy to share our article just published in Brain. Well done Dr. Saira Mirza and Maurice Pasternak. @GENFI1 @Sunnybrook @UofT_DoM @UofTNeurology : Disease-modifying effects of TMEM106B in genetic frontotemporal dementia: a longitudinal GENFI study
academic.oup.com
Mirza et al. show that a variant in the TMEM106B gene confers protection against frontotemporal dementia (FTD) by modifying the effects of FTD mutations—in
0
8
20
I'm happy to share that our paper describing our novel TMEM106B overexpression mouse model is now out in Molecular Neurodegeneration 🧠 It’s been a project with highs and lows, and seeing it published is incredibly rewarding. Thanks to everyone who was part of the journey 🙌.
'Increased #TMEM106B levels lead to #lysosomal dysfunction which affects synaptic signaling and neuronal health' Jolien Perneel, Miranda Lastra Osua, Sara Alidadiani... Xiaolai Zhou & Rosa Rademakers @RademakersRosa @CMN_VIB @UAntwerpen
https://t.co/h6fdnzCHjo
3
5
40
We are proud to share our latest Genetic Information Guide, on the GRN variants associated with FTD. Focused on concepts of most interest to, and written to be accessible for, those impacted. https://t.co/lITE9XmK24
endthelegacy.org
0
3
6
Mis-spliced transcripts generate de novo proteins in TDP-43-related ALS/FTD | Science Translational Medicine
science.org
Loss of TDP-43 function results in the expression of de novo proteins from mis-spliced mRNA transcripts.
0
10
52
Study identifies a fluid biomarker of TDP-43 dysfunction, a central pathological feature of the #ALS-FTD disease spectrum, and demonstrates that such loss of TDP-43 splicing repression occurs presymptomatically @JHUPath, @HopkinsNeuro @Katie_E_Irwin
https://t.co/L54ow56MSm
1
34
121
Very excited for my first time at #SfN23! 🧠 Find me at my poster tonight at the TPDA poster session (KK9) - or come check out my talk (NANO53.02) on Tuesday morning. Special thanks to SfN-Gatsby Charitable Foundation for the support!
0
1
19
This #FTDAwarenessWeek, we're proud to be advocating for FTD research. @RademakersRosa and her team are working hard to advance research on FTD 👇 To learn more about their work, visit 🔗 https://t.co/v1Ygajwwkd
#endFTD #WeCareAboutFTD 🩵
0
7
20
Preprint: Control of mitophagy initiation and progression by the TBK1 adaptors NAP1 and SINTBAD We are excited to share our latest study on how PINK1/Parkin mitophagy initiation and progression is regulated. Out now on @biorxivpreprint! 1/ https://t.co/xjxLZvd8eL
biorxiv.org
Mitophagy preserves overall mitochondrial fitness by selectively targeting damaged mitochondria for degradation. The regulatory mechanisms that prevent PINK1/Parkin-dependent mitophagy and other...
3
16
38
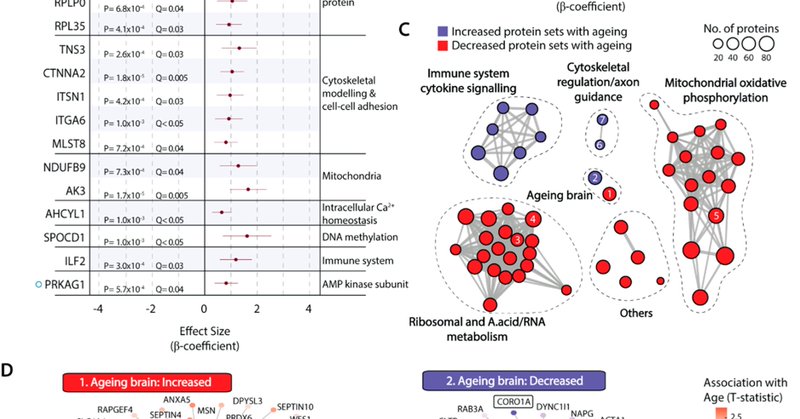
My first first-author publication about #TMEM106B in the #hippocampus of ageing cognitively normal individuals has just been published @MolNeuro
https://t.co/v4vVuAqrmn
#proteomics #lipidomics #neurodegeneration
link.springer.com
Molecular Neurodegeneration - The risk for dementia increases exponentially from the seventh decade of life. Identifying and understanding the biochemical changes that sensitize the ageing brain to...
1
6
18
'The major #TMEM106B dementia risk allele affects TMEM106B protein levels, fibril formation, and myelin lipid homeostasis in the #ageing human hippocampus' @jun_lee4 ... Anthony S Don @CPC_usyd @Sydney_Uni
#lipidomics #proteomics
https://t.co/nvulTeyGXK
0
4
6
Here it is!! 🤩 Excited to announce that my first first-author paper from my PhD is now available as a preprint! Many thanks to my lab @VIB_Switch_Lab and our collaborators for the effort!
N-glycosylation as a eukaryotic protective mechanism against protein aggregation https://t.co/pohOjn3P2n
#bioRxiv
0
4
34
Beaten to tweeting our paper by @ShorterLab! Check out the RNAs that bind C9orf72 polyPR -we hope a useful resource for the field @LSAjournal
life-science-alliance.org
An intronic GGGGCC repeat expansion in C9orf72 is a common genetic cause of amyotrophic lateral sclerosis and frontotemporal dementia. The repeats are transcribed in both sense and antisense direct...
Transcriptome-wide RNA binding analysis of C9orf72 poly(PR) dipeptides, delighted for this work with @isaacs_adrian, @RubikaBalendra, @PataniLab, @ule_lab et al. to be out in @LSAjournal!
2
7
46