Jingyi Wei
@JingyiWei4
Followers
430
Following
1K
Media
13
Statuses
370
Postdoc at MIT BCS, Guoping Feng lab, Phd @Stanford Bioenginnering, Konermann lab @SKonermann, interested in brain disorders, CRISPR screens, and gene editing
Joined January 2020
RT @pdhsu: We release 300 million single cells (and counting) for virtual cell modeling, our first AI agent work @arcinstitute, and 100 mil….
0
60
0
RT @EricTopol: Just another >300 million single cells, agentic #AI life science, large language of life models, perturbations, open-access….
0
102
0
RT @SKonermann: Excited to be launching this resource @arcinstitute for the scientific community today with the first 300M cells, including….
0
15
0
RT @idmjky: Thrilled to share that EVOLVEpro is now published @ScienceMagazine Since our preprint, we now demonstrate low-N eningeering of….
www.science.org
Directed protein evolution is central to biomedical applications but faces challenges such as experimental complexity, inefficient multiproperty optimization, and local maxima traps. Although in...
0
81
0
RT @pdhsu: Delighted to see @arcinstitute research highlighted by Vox's Future Perfect 50 list!.
0
5
0
RT @HowardYChang: I am excited to join storied biotech pioneer @Amgen as Chief Scientific Officer and Head of Research. I look forward to p….
www.amgen.com
0
39
0
RT @pdhsu: Today we report Evo, a generalist foundation model for biology, on the cover of @ScienceMagazine. Trained on billions of DNA nuc….
0
310
0
Our DjCas13d enzyme and guide backbones are on Addgene now! CRISPR and RNA lovers, try it out! 😊.
Excited to share my major PhD work: .(right before my defense and critical time for my postdoc search😊) Summary: an accurate CNN model for Cas13d guide efficiency prediction + a new non-toxic and highly specific enzyme (DjCas13d) for RNA targeting!.
0
3
18
Great work and very comprehensive study! Happy to test in hESCs (and it worked super well!). Give it a try! 😉.
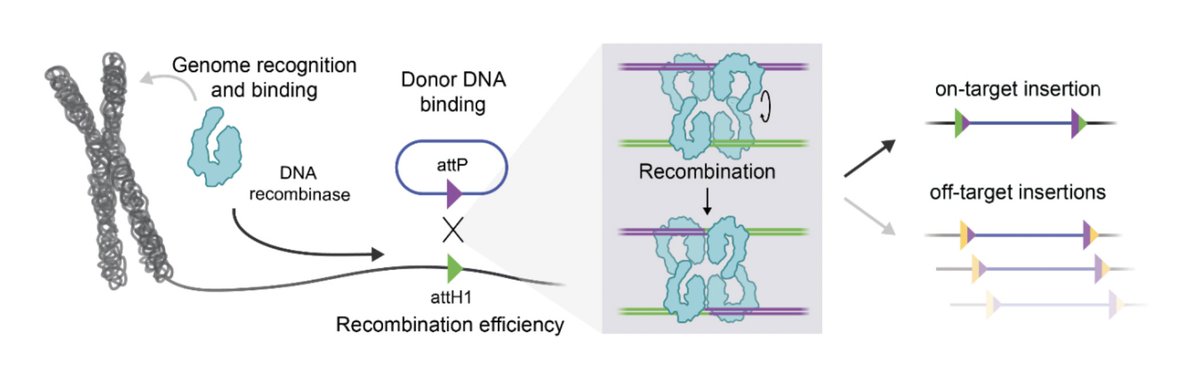
🧵In new work, we report a systematic engineering roadmap to optimize large serine recombinases (LSRs) for direct, site-specific insertion into the human genome 🧬. We achieved over 50% insertion efficiency and 97% genome-wide specificity, a 10X improvement over our previous work
0
0
12
RT @GallowayLabMIT: Levels of gene expression matter! So how can you control a transgene precisely? Is there a tool that can ensure uniform….
0
129
0
RT @FelixHorns: I’m beyond excited to launch my lab today and to join the amazing communities at @arcinstitute and @Stanford Genetics!.
0
17
0
RT @arcinstitute: It’s not often we catch all of our Core Investigators in one place, standing still, but we did it! Welcome @FelixHorns -….
0
10
0
RT @thenormanlab: Excited to share our preprint "Multiome Perturb-seq unlocks scalable discovery of integrated perturbation effects on the….
0
86
0
RT @pdhsu: Please RT! @BrianHie and I are hiring joint @arcinstitute postdocs to work on ML in biology. Projects include reimagining synthe….
0
83
0
RT @SKonermann: I'm excited for this fundamental new RNA-guided mechanism out of @arcinstitute to come out today in back-to-back papers in….
0
13
0
Just graduated from Stanford and headed to MIT BCS (Guoping Feng lab) for postdoc research!.Thanks to my PI @SKonermann, teachers, family and friends for your love and support!.Excited to embark on a new journey! (Surprised and proud to see our Cas13d in the MIT museum! 🤣
0
4
61
RT @GogoLiu: We look forward to working with our scientific colleagues to optimize and expand the application pote….
www.science.org
Bacterial GII-C intron ribozymes are programmable and self-guided endonucleases active for sequence-specific DNA manipulation.
0
38
0
RT @cyrus_tau: .@sitaritapita and I are excited to share RESPLICE, a new RNA-programmable mode of editing that leverages RNA targeting CRIS….
www.biorxiv.org
Splicing bridges the gap between static DNA sequence and the diverse and dynamic set of protein products that execute a gene’s biological functions. While exon skipping technologies enable influence...
0
37
0