Javier Mendoza-Revilla
@JavierMenRev
Followers
770
Following
1K
Media
26
Statuses
581
Computational Geneticist @instadeepai and adjunct lecturer at @CayetanoHeredia. Previously at @UGI_at_UCL🇬🇧 and @institutpasteur🇫🇷. Views my own.
Paris, France
Joined February 2013
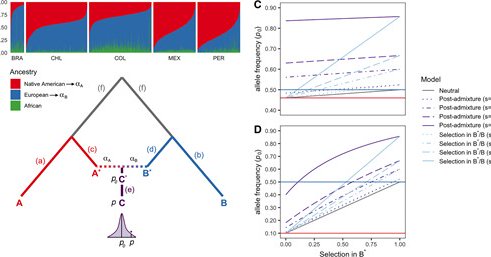
Our study with @g_hellenthal on detecting adaptive signals before and after European contact in #admixed Latin Americans is finally out in @MolBioEvol
https://t.co/MaMBgZutOa
academic.oup.com
Abstract. Throughout human evolutionary history, large-scale migrations have led to intermixing (i.e., admixture) between previously separated human groups
2
22
62
Nucleotide Transformer is a series of genomics foundation models of different parameter sizes and training datasets which can be applied to various downstream tasks by fine-tuning. @instadeepai
https://t.co/R31y1UNU6w
1
30
88
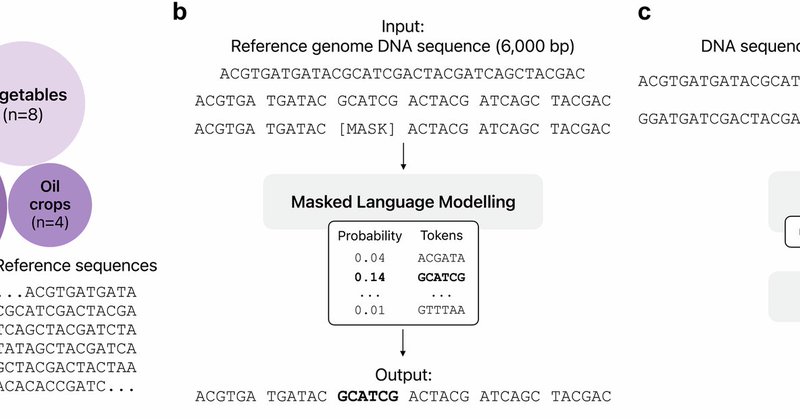
We were invited to write a review article on DNA/genomic language models (gLMs). We took this occasion to gather our thoughts on promising applications, and major considerations for developing and evaluating gLMs. Pls share with your colleagues: Preprint:
arxiv.org
Large language models (LLMs) are having transformative impacts across a wide range of scientific fields, particularly in the biomedical sciences. Just as the goal of Natural Language Processing is...
1
109
367
A DNA-based large language model, AgroNT, trained on multiple plant genomes, can accurately predict various molecular phenotypes within plant species, including important crops. @instadeepai @JavierMenRev @thomas_pierrot @LopezMarie05
https://t.co/K1zehDejL7
nature.com
Communications Biology - A DNA-based large language model, AgroNT, trained on multiple plant genomes, can accurately predict various molecular phenotypes within plant species, including important...
0
5
8
Our work on the development of a #DNA Large Language Model trained on genomes from various plants, #AgroNT, is now published in @CommsBio
https://t.co/GKWTUT9Btw
nature.com
Communications Biology - A DNA-based large language model, AgroNT, trained on multiple plant genomes, can accurately predict various molecular phenotypes within plant species, including important...
0
0
8
ChatNT: A Multimodal Conversational Agent for DNA, RNA and Protein Tasks https://t.co/rEEvFqnlaE
1
23
75
Very proud to announce ChatNT, the first multimodal conversational agent for biological sequences🧬. ChatNT can be simply prompted with a question and a nucleotide sequence to solve DNA, RNA and Protein tasks 🧑🔬! 📚Paper: https://t.co/mU6HsNJweA 🌐Blog:
instadeep.com
The human genome, containing the entirety of our genetic blueprint, holds the keys to understand the intricate workings of our cells and bodies. How our cells respond to signals, build proteins, and...
The NT Family is growing! 🐣 ✨ Introducing ChatNT, a Conversational Agent designed to analyse genomics sequences and address a wide range of key biological questions, assisting scientists in their daily work 👩🔬 📚Paper: https://t.co/gkHiEgN8rb 🌐Blog: https://t.co/ods98i8s9K
1
8
36
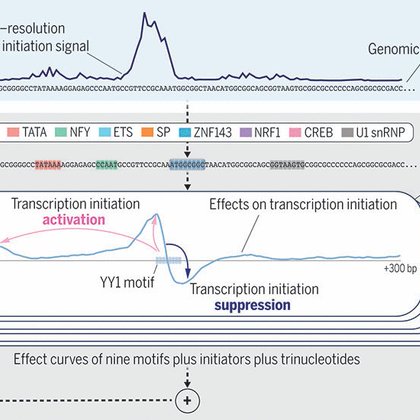
Glad that our sequence model of promoters in human genome is now published in @ScienceMagazine. Check out the paper for a deep dive into the sequence basis of transcription initiation at the basepair level:
science.org
Transcription initiation is a process that is essential to ensuring the proper function of any gene, yet we still lack a unified understanding of sequence patterns and rules that explain most...
14
80
323
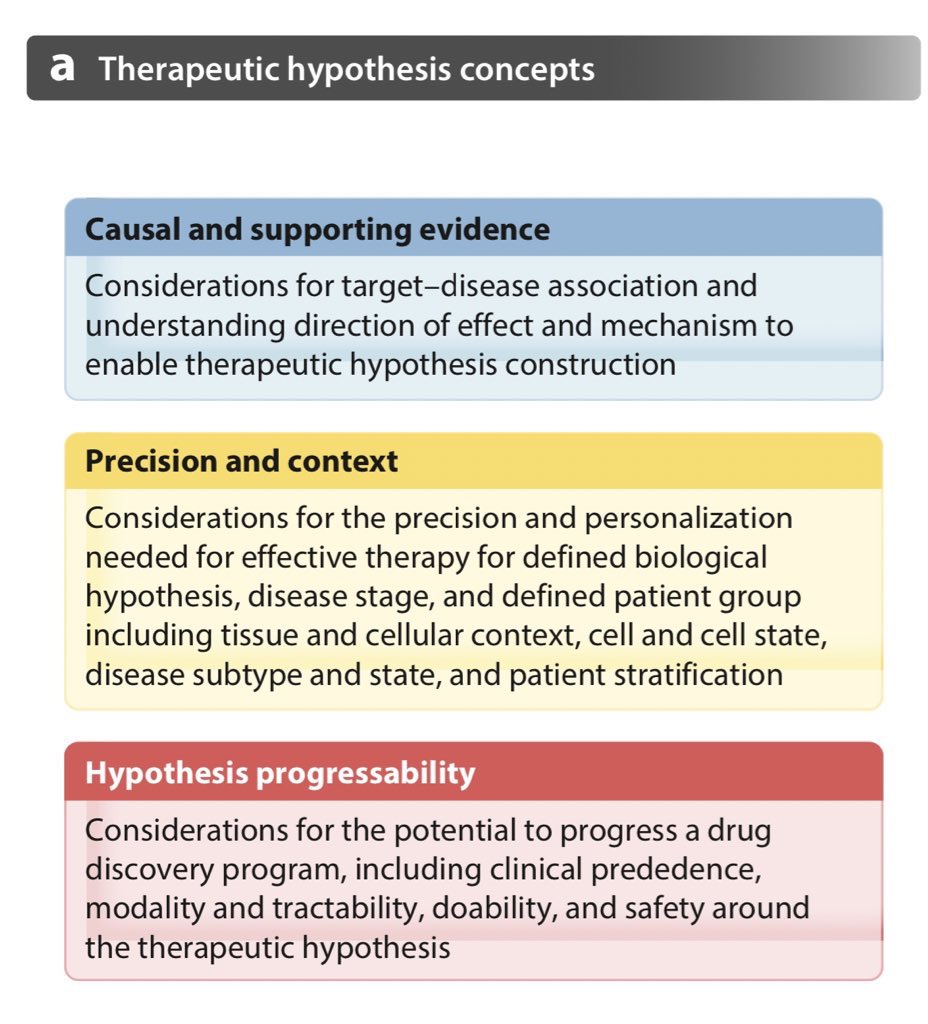
Out now! Together with members of our Exec & Science Leadership Team @emblebi @sangerinstitute @GSK @genentech @pfizer @bmsnews @sanofi we share our perspective on how @OpenTargets we use human genetics & genomics for drug target ID & prioritisation: https://t.co/i72vMOQbua
0
12
32
✨ Meet SegmentNT: the first-ever LLM capable of annotating DNA sequences at single nucleotide resolution. Built on top of our Nucleotide Transformer, SegmentNT offers precise genome annotation surpassing traditional methods and can offer deeper insights into our genome 🧬
2
25
60
InstaDeep's @JavierMenRev will share how AI systems - and LLMs specifically - deepen our understanding of the Maize genome tomorrow at 7:20 pm at the Maize Genetics Meeting in Raleigh, NC. #MGM2024 Read the paper: https://t.co/1832A5M5eB
0
1
9
Reposting this criticism of All of Us and their use of ancestry labels https://t.co/H476c45O8x
The analyses highlighted by the authors risk exacerbating the confusion. Genetic ancestry does not map neatly onto, nor is it highly concordant with these geographic and ethnic categories. 5/n
3
27
104
We released the weights and downstream task datasets of the agroNT, our 1B parameters DNA foundation model for plant genomics 🧬🪴. Check out our github ( https://t.co/oyMSkhXf8Q) for Jax lovers⚡❤️and our @huggingface space ( https://t.co/z4uYMEec0y) for the pytorch version🤗.
huggingface.co
1
52
141
I am looking for PhD research interns at the @instadeepai Paris office for the coming summer. Join us if you are interested in DNA LLMs and regulatory genomics! We have different projects available and are flexible to find one that fits your PhD. Link:
1
16
39
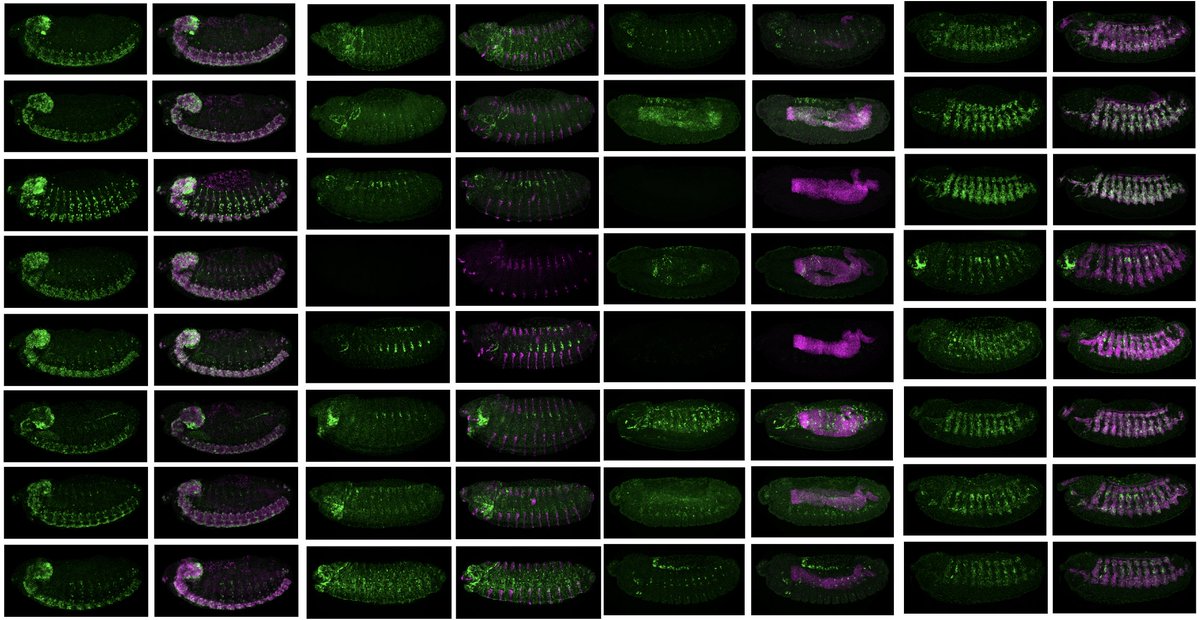
We are very excited to present a major breakthrough achievement – the de novo design of synthetic enhancers for selected tissues in fruit fly embryos in vivo using deep- and transfer learning, @deAlmeida_BPet al published today in @Nature
https://t.co/TIlPrBKYaK. Thread 👇(1/N)
21
199
672
If you are interested in gene-environment interactions, molecular phenotypes and single-cell genomics, join us at @institutpasteur in the sunny Paris! Post-doctoral position in single-cell genomics at Institut Pasteur in Paris
0
30
32
(2) GWAS Transferability: PRS show much greater accuracy in individuals of European ancestry than in others. Thus, the PRS they report may be highly inaccurate for many individuals (admixed/from underrepresented populations) and potentially even irrelevant for some.
1
0
7
(1) Pleiotropy: Meaning, a variant at a locus can affect multiple traits simultaneously. Also, pleiotropy can be antagonistic, implying that a variant may be simultaneously harmful AND beneficial. What, then, will these types of screenings be selecting for?
1
0
7
Putting aside the ethical concerns related to PRS in embryos, two major drawbacks of this screening approach quickly come to mind:
1
0
1
I am very interested to know what other population and statistical #geneticists think about this.
We’re excited to announce the first whole genome screening for embryos is available! Parents can get 100x more data about their embryos’ genomes, empowering them to make an informed decision and give their baby the best chance at a healthy start.
6
1
24