Zhicheng Cui | 崔志成
@Chen3dem
Followers
395
Following
528
Media
6
Statuses
375
Ph.D. @TAMUBCBP , Postdoc @TheHurleyLab Bluesky:https://t.co/fuHMA16lgu
Berkeley, CA
Joined July 2013
Thrilled to present the structure of mTORC1 on membrane. We are at an exciting time of using SPA to resolve high-resolution structures of complexes that land on the surface of membranes! Looking forward to seeing what can be done for this research frontier in the near future!
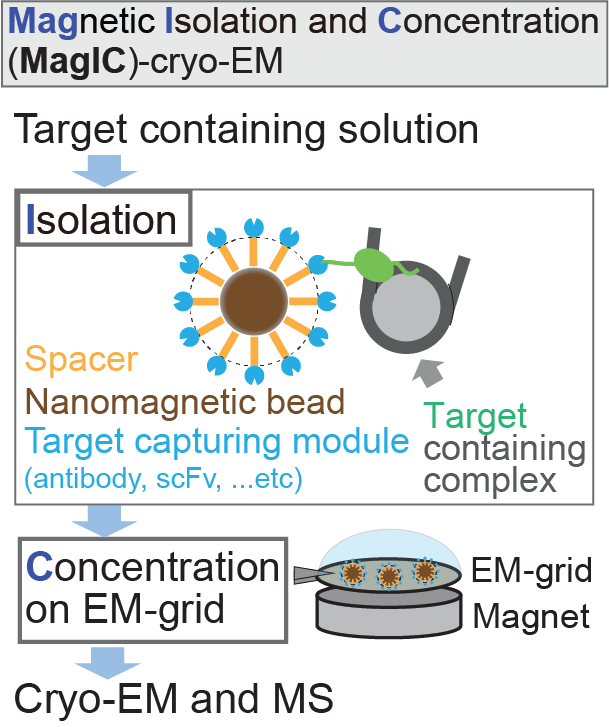
Proud of new work https://t.co/VVD4eyFOlc from the lab by Zhicheng (Chen) Cui @Chen3dem using single-particle cryo-EM on liposomes to show how mTORC1 integrates nutrient and growth factor signaling at lysosomes, collab. w/ @G__Napolitano, A. Esposito, and A. Ballabio.
4
14
91
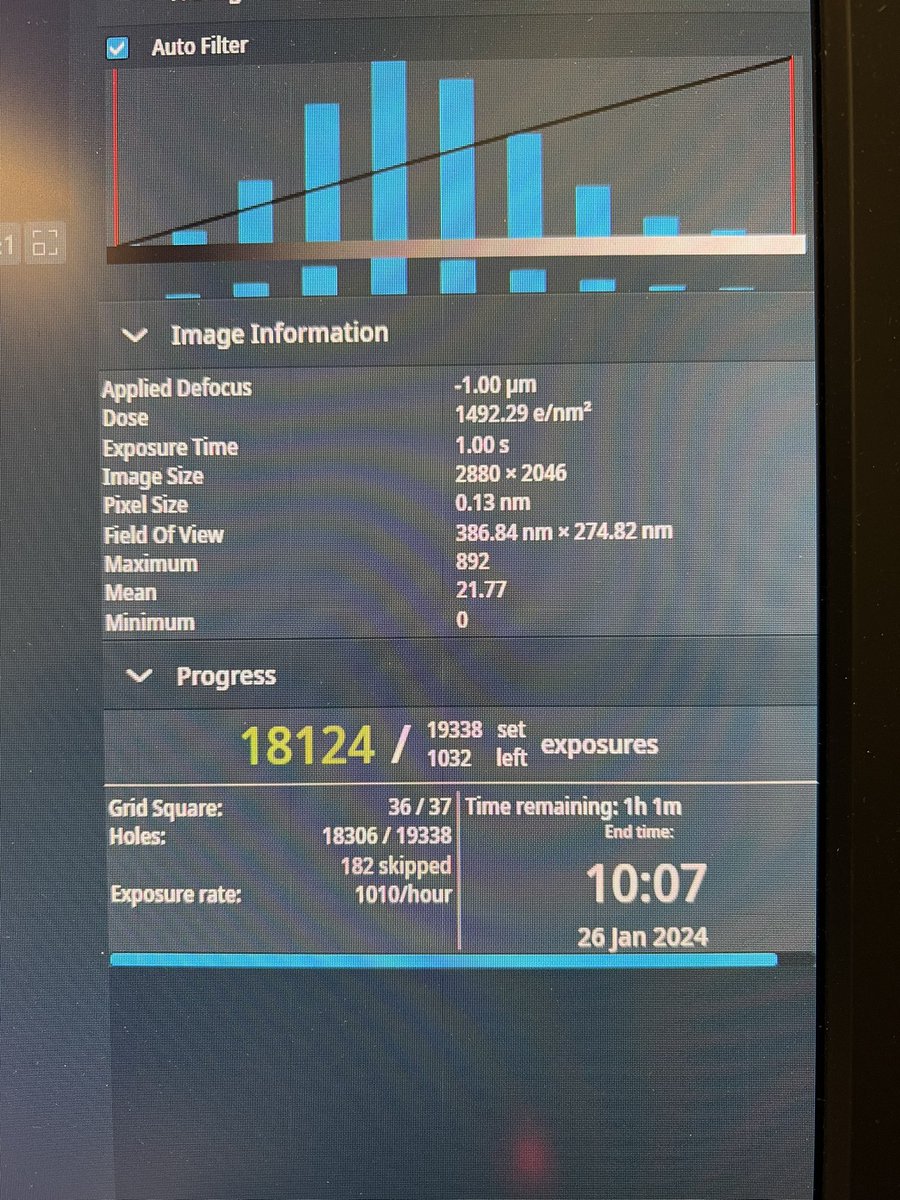
Defining the EM-signature of successful cell-transfection https://t.co/88dEYO9Xpj
#biorxiv_cellbio
0
17
98
I am thrilled to share the first preprint from my lab at @Mizzou_Biochem! My colleague @CKeithCassidy and I determined the structure of apoB100 from human low-density lipoprotein (LDL). This protein is wild! https://t.co/uh5HZRMd2b
4
23
102
Check out our new publication on the fate of cGAS in cycling cells! https://t.co/hwwgXb0w0G It has been established that cGAS enters the nucleus and tethers to nucleosomes during mitosis. However, what is the ultimate fate of nuclear cGAS? 1/n
5
35
117
Check out the 2nd SnapShot of the ribosome biogenesis series featuring the 60S ribosomal subunit! This was a great collaboration w Ed Hurt & Roland Beckmann. Read the SnapShot & watch the animation w narration here: https://t.co/qp5R4sLXyL
8
75
248
🎉 We are thrilled to announce the release of the latest structure of the #cGAS complex in PDB as https://t.co/WpAGGpgm9b🌐✨Stay tuned for the full story in our upcoming paper, where we unveil the secrets behind cGAS transportation and its ultimate destiny. #Immunity
4
34
182
Happy to share the publication of our paper in @Nature Conformational ensembles of the human intrinsically disordered proteome Work led by @GiulioTesei and @AnnaIdaTrolle. I'll post more later, but for now here is a link: https://t.co/b3aZ6BbGpB and a short movie about the work
50
287
1K
I'm happy to share ColabFold-Pipeline-Toolkit, an easy recipe book put together to help streamline the use of ColabFold for newcomers without a computing background who want to perform high-throughput screens with ColabFold BATCH. 🧵 https://t.co/QrST6D0laR
github.com
The aim of this repository is to provide simple tools to help those working with ColabFold BATCH both for pre and post-processing steps. - andyposbe/ColabFold-Pipeline-Toolkit
3
95
239
Wrists sore from pipetting? Check out our preprint on “Surface Patterned Omniphobic Tiles (SPOTs): a versatile platform for scalable liquid handling”. https://t.co/hso6nugEFN
2
22
63
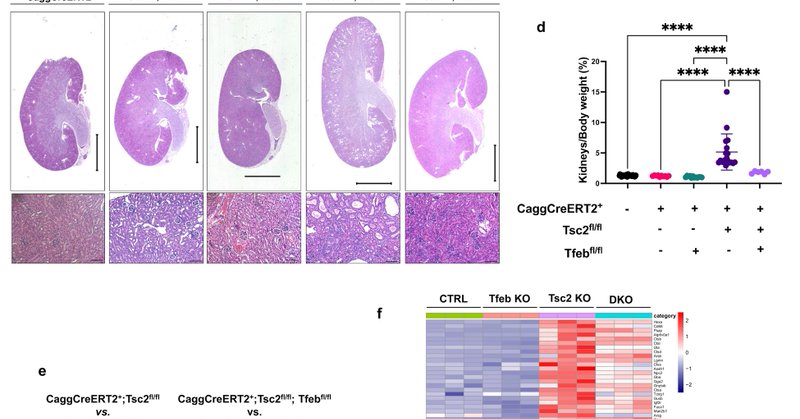
New paper from Alesi (@NicolaAlesi) et al. where the authors show that TFEB is the primary driver of renal disease and mTORC1 hyperactivation in Tuberous Sclerosis Complex (@LisaHenske). https://t.co/6nXz5wFa8e
nature.com
Nature Communications - Tuberous Sclerosis Complex (TSC) is caused by TSC1 or TSC2 mutations, leading to hyperactivation of mechanistic target of rapamycin complex 1 (mTORC1) and tumors in multiple...
0
2
14
Macrocyclic Peptide Drugs From the Ground Up | Science | AAAS
science.org
Macrocyclic Peptide Drugs From the Ground Up
0
1
2
The Shin lab is open for business on the 12th floor @CRI_UTSW!! Come chat about science, life in Dallas or anything😎
10
23
145
Best advice I ever got for giving a talk, from my PhD supervisor, repeated to the lab today. (Dolled up by a TIDA UP state on the right.)
3
35
282
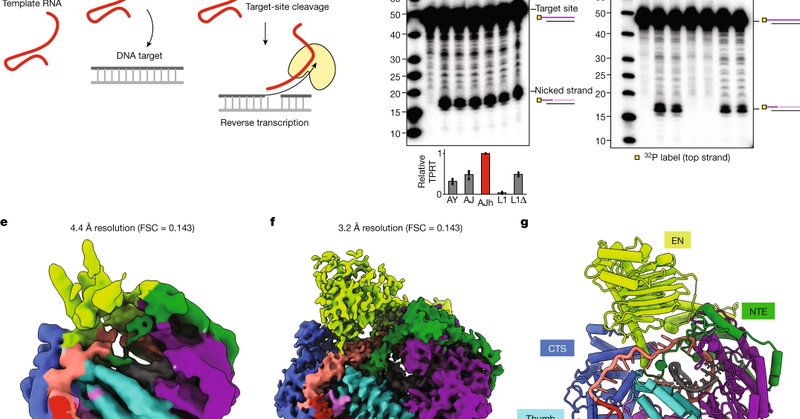
🔥🔥How does the LINE-1 retrotransposon jump around in the human genome? Super excited to present my postdoctoral work answering this published today @Nature together with twitterless Kathy Collins's lab and @NogalesLab @UCBerkeley
https://t.co/quoQOpZb5X A 🧵
nature.com
Nature - Human LINE-1 ORF2p relies on upstream single-stranded target DNA to position the adjacent duplex in the endonuclease active site for nicking of the longer DNA strand, with a single nick...
11
89
409
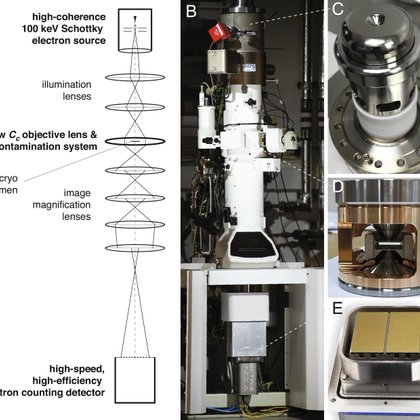
Happy to have played a small part in this and we're still working on a few things to push this further. https://t.co/8M4hIVRaKR
pnas.org
Electron cryomicroscopy can, in principle, determine the structures of most biological molecules but is currently limited by access, specimen prepa...
5
8
49
Google just launched Duet AI, and it might just overtake ChatGPT and Microsoft Copilot! Here are 5 mind-blowing features you don't want to miss:
153
1K
6K
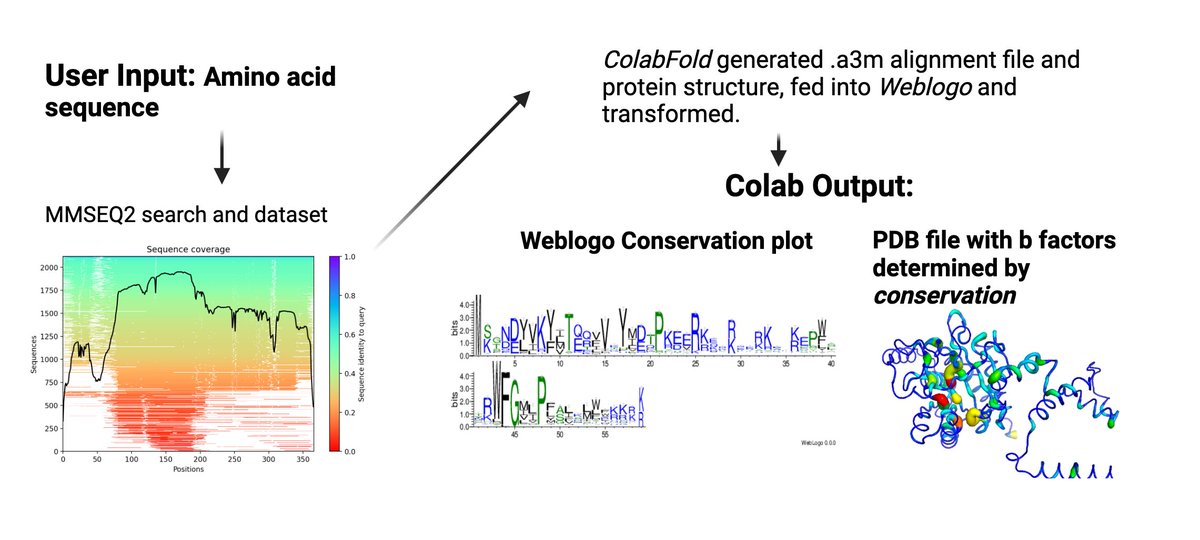
We made a little conservation of amino acids tool, combining Weblogo and ColabFold, which you can run in the browser with a single sequence. Since some major conservation servers are down. Hope it's useful! https://t.co/DSe477mUyr
@CRodriguesLab @pstansfeld @sokrypton
8
78
318