Fran Supek
@FranSupek
Followers
3K
Following
9K
Media
95
Statuses
689
🧬Genetics geek,🤖AI enthusiast. PI @GenomeDataLab studying DNA repair, mutations, epigenome, NMD, cancer. Prof @UCPH_Research, GL @IRBBarcelona 🎲Chaotic good
Copenhagen/Barcelona
Joined July 2009
Thanks to organizers for inviting!
S2.2. DNA break repair in the CRISPR era @EMBO 📢 Using combinatorial CRISPR/Cas12 edits and mutational signatures to study genetic interactions in DNA repair pathways - Fran Supek @FranSupek #47congresosebbm
0
1
7
📣We’re recruiting 1-2 Group Leaders! @NCMBMnews is looking for early-career researchers ready to establish their independent research groups! 🧬With an attractive start-up package, help us shape the future of molecular medicine in 🇳🇴 and be part of the @NordicEMBL! @embl
0
5
12
Congrats to our CRISPy fresh doctor 👏👏 Marcel McCullough for successfully defending his PhD thesis: 🧬✂️Combinatorial Gene Editing in Human Cells to Model Deficiencies of DNA Repair in Cancer Thx to committee members @CeronLab @LValleResearch and @DSzuts for great discussion!
1
3
8
Copenhagen is on a roll. It was ranked the safest major city in the world last year too. The food scene is sublime. Public transit phenomenal. So livable, so family friendly. Can't buy this mix of attributes for any amount of money in the US.
131
136
3K
We observed that tRNA genes mutate like no tomorrow 👇
In this fascinating preprint, the authors uncover an eye-opening feature of tRNA genes: they’re insanely prone to mutation. Analyzing somatic mutations in cancer tissues, they report that tRNA genes are up to 9 times more mutated than protein-coding genes. Interestingly, the
0
4
20
Join us at BRIC/Finsen -- start your lab at the University of Copenhagen!
WE ARE RECRUITING: A tenure-track group leader position in translational cancer research is available at the Finsen Laboratory in Copenhagen. Come join an excellent and friendly research environment combining both hospital and university affiliated groups https://t.co/K9ebKtLysf
0
1
1
Why care? This gives: -new resistance biomarkers (eg MAP2K4 in breast) -framework to compare pre/post-treatment genomes -warning about "driver genes" that are evol. relics Huge thanks to funders & bravo @Ahmed_IS_Khalil👏 ➡️Nice @IRBBarcelona write-up:
irbbarcelona.org
Just as species adapt over generations, our body’s cells accumulate DNA changes throughout life. Most are harmless, yet a few “driver” mutations give a cell a competitive edge and can spark cancer....
1
0
2
Bonus analysis: "cancer drivers" that are under selection in healthy tissues🧬. NOTCH1 (known) and ARID1A mutations promote clonal expansions w/o cancer transformation. Challenges assumptions about what makes a true driver - maybe some mutations are just along for the ride 4/5
1
0
1
Found 11 genes under therapy-associated selection - some expected (ESR1, EGFR) but surprises too: 💊PIK3CA selected in antiestrogen-treated 💊SMAD4 under pressure in 5-FU-treated 💊STK11 mutations with methotrexate Resistance often piggybacks onto existing oncogenic programs 3/5
1
0
1
Chemo changes mutation rates AND selection pressures - a confounding mess.🔥 DiffInvex cuts through this by using non-coding DNA as evolutionary baseline, separating signal (driver mutations) from noise (therapy-induced mutagenesis) in ~30 cancer types: 🔗 https://t.co/jITZB7oapr
nature.com
Nature Communications - Identifying mutations in cancer that are causal for resistance to chemotherapy remains challenging. Here, the authors develop DiffInvex, a computational framework to...
1
0
1
🧵Ahmed’s @GenomeDataLab paper on evolution of chemotherapy resistance is out in @NatureComms! We developed DiffInvex - a method tracking how #cancer driver genes change under therapy, applied to ~10k genomes. Key insight: resistance often emerges through existing pathways. 1/5
2
5
12
Meanwhile in Europe: the Impossible Burger is still 100% illegal. Thank you, anti-GMO people, for protecting us from sustainable, humane and delicious plant-based burgers. 🙃
65
116
1K
Grateful for the award of this collaborative project with Claus Sørensen group at BRIC, who will apply their awesome #CRISPR-Select method (Niu et al 2022 Nature Genetics) to introduce heterozygous edits into human cells. ➡️ https://t.co/IRq52LY2Ac
dff.dk
52 talentfulde forskere skal med deres nyskabende og excellente forskningsprojekter løfte dansk forskning til nye højder. Danmarks Frie Forskningsfond har netop uddelt 318 millioner kroner til...
0
0
0
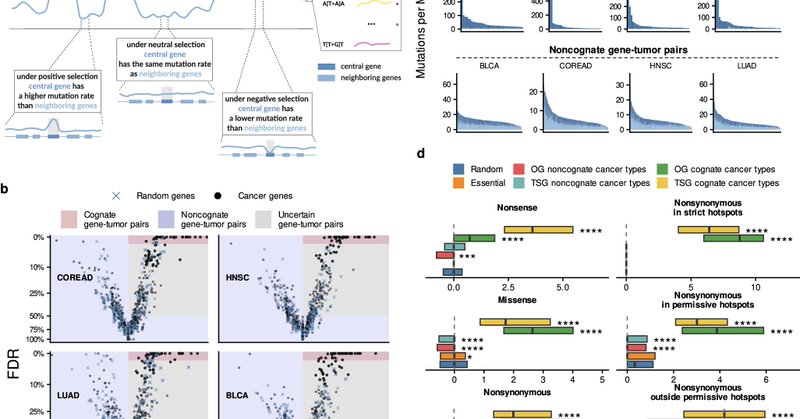
🧬 We aim to identify TSG #mutations that could be #targets for therapeutic inhibition - an idea supported by our recent finding that tumor suppressor #genes, paradoxically, can get copy number gains that drive cancer (Besedina & Supek 2024 Nature Comms https://t.co/k9Sj37pkrD)
nature.com
Nature Communications - Inferring the emergence and selection of cancer drivers remains a daunting task. Here, the authors develop MutMatch, a statistical method to analyse somatic mutation rates...
1
0
2
Come and join us! We’re hiring a new Group Leader in Generative Biology at the @sangerinstitute Building AI models or the data to train them? Core funding of >$130M a year for a faculty of ~30. pls RT! https://t.co/skA0MAOTyZ
https://t.co/cZ8i3TigJH
1
27
61
🚨 We have extended the deadline for submissions! 🚨 📅 New deadline: Feb 13, 2025, AoE - so you can finish just in time for Valentine's day 🥰 🔗 More details on our website (link in bio)
0
7
10
We are organising https://t.co/bmP3bmhJHq at#ICLR2025. Topics: RNA, DNA and cell LLMs, structure, modifications, correction, variant calling... Deadline: Feb 10th 2025 Time for revision: 1 month Accepted papers - the opportunity to be invited by Nature Methods for submission
ai4na-workshop.github.io
AI4NA Workshop @ ICLR'25
0
8
19
Double PhD defense day in the lab 🤩 Congrats to our dr. Guille Palou @gpaloumarquez, fresh from the oven 🎊🥳 and similarly so our newly-minted dr. Ignasi Toledano @IgnasiToledano! Amazing defense both, bravo 👏👏
1
4
11
🧬🤖By bringing together machine learning & biology, we will explore how AI can address key challenges in nucleic acids research, such as structure prediction, understanding RNA/DNA interactions, and designing therapeutic molecules Our call for papers: ➡️ https://t.co/spCGf5hIAK
ai4na-workshop.github.io
We invite submissions to the 1st AI for Nucleic Acids Workshop at ICLR 2025.
0
0
1