Ziyu Lu
@ziyu__lu
Followers
112
Following
149
Media
0
Statuses
19
PhDing @RockefellerUniv @junyue_cao Lab
New York
Joined May 2019
Thrilled to share our latest preprint led by the fantastic student Ziyu @ziyu__lu! We constructed a scATAC-seq atlas profiling > 500 mouse tissues to explore cell population dynamics during mammalian aging and the associated chromatin landscape changes: https://t.co/AiIMf5r1hV
2
26
104
I'm thrilled to share our new #bioRxiv preprint from @junyue_cao lab at @RockefellerUniv! We present PerturbFate, a high-throughput single-cell CRISPR platform profiling nascent/pre-existing RNA, steady-state RNA & chromatin accessibility.
biorxiv.org
High-throughput genomic studies have uncovered associations between diverse genetic alterations and disease phenotypes; however, elucidating how perturbations in functionally disparate genes give...
8
42
306
Thrilled to introduce IRISeq, a scalable, cost-effective and imaging-free spatial genomic technique for mapping mammalian brain aging, developed by incredible Abdul @Abdul_Squared and Weirong from our lab at @RockefellerUniv! https://t.co/rvafkbYecP
3
51
153
Wei Zhou (@Wei_Zhou_1989) is a staff scientist in @junyue_cao's lab. She studies how a cell population maintains homeostasis, and how it is disrupted in aging and aging-related diseases, through profiling + perturbing cell dynamics at single-cell resolution. #AANHPIHeritageMonth
0
8
22
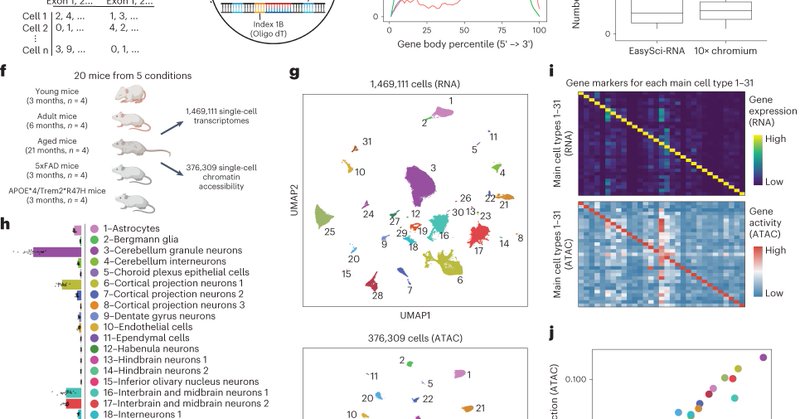
EasySci is now published! 🌟 A big congratulations to Andras, @ziyu__lu, and Jasper from our team at @RockefellerUniv. Planning to prepare a scRNA-seq library for 1 million cells on a budget of just $700?Discover our detailed protocol here:
nature.com
Nature Genetics - Single-cell transcriptomes and single-cell chromatin accessibility profiles generated using EasySci provide a global view of aging and Alzheimer’s pathogenesis-associated...
7
109
398
Single-cell characterization of many genetic mutants at the scale of the entire organism! A great collaboration with @malte_spielmann lab and my fantastic colleagues in @JShendure lab!
So happy to share the final version of our single cell whole embryo paper out today @Nature ! Such a great collaboration with @junyue_cao & @JShendure labs @UKSH_KI_HL @UniLuebeck @MPI_MolGen @kieluni Check out the summary by @HenckJana :
0
4
26
In the latest issue! Tracking cell-type-specific temporal dynamics in human and mouse brains
1
27
92
How does aging impact the proliferation and differentiation rates across diverse cell types in the brain? Explore this question with TrackerSci in @CellCellPress: https://t.co/t0nShT9lLK. Kudos to the incredible @ziyu__lu and Melissa Zhang of our lab @RockefellerUniv!
6
32
125
PerturbSci-Kinetics for high-throughput functional characterization of RNA dynamic regulators is published today @NatureBiotech, led by fantastic @xu_zxu from our lab @RockefellerUniv!
Dissecting key regulators of transcriptome kinetics through scalable single-cell RNA profiling of pooled CRISPR screens https://t.co/QIrbZioqKg
18
59
258
Dissecting key regulators of transcriptome kinetics through scalable single-cell RNA profiling of pooled CRISPR screens https://t.co/QIrbZioqKg
0
64
214
Our next @cegs_ica virtual seminar will be on 4/4 at 12PM ET @xu_zxu will present 'PerturbSci-Kinetics' a novel combinatorial indexing method to capture single-cell transcriptome, nascent RNA, and sgRNA identity in pooled CRISPR screens. Free registration: https://t.co/Sk1p9vFadv
0
15
52
Retrospective analysis of enhancer activity and transcriptome history https://t.co/r2TtTYLvFl
4
38
130
Congrats @xu_zxu This is brilliant!!
I am thrilled to share our latest research, PerturbSci-Kinetics, a novel low-cost and scalable combinatorial-indexing single-cell technique enabling the co-capture of transcriptomes, nascent transcriptomes, and genetic perturbations. (1/5)
1
0
1
I am thrilled to share our latest research, PerturbSci-Kinetics, a novel low-cost and scalable combinatorial-indexing single-cell technique enabling the co-capture of transcriptomes, nascent transcriptomes, and genetic perturbations. (1/5)
Excited to share our latest preprint from Cao Lab - PerturbSci-Kinetics, led by brilliant @xu_zxu! The technique captures single-cell transcriptome, nascent RNA, and sgRNA identity in pooled CRISPR screens, for identifying key regulators of RNA kinetics.
2
7
45
Excited to share our latest preprint from Cao Lab - PerturbSci-Kinetics, led by brilliant @xu_zxu! The technique captures single-cell transcriptome, nascent RNA, and sgRNA identity in pooled CRISPR screens, for identifying key regulators of RNA kinetics.
7
41
161
Interested in massive-scale combinatorial indexing technologies? Check out our next @cegs_ica virtual series. Two students from @junyue_cao's lab will introduce EasySci-RNA (1M cells for $700!) to study aging and neurodegeneration Free registration: https://t.co/tfBwJIOkBl
0
22
74
Really excited to have my first preprints out!! Huge thanks to the exceptional mentor @junyue_cao, all lab members, and support from @RockefellerUniv !!!!
Hello World! Thrilled to share the first two preprints from Cao lab at @RockefellerUniv. Both focus on understanding cell population dynamics in aging/AD through new single-cell genomic techniques, led by our fantastic @ziyu and Andras!
0
4
16
A global view of aging and Alzheimer's pathogenesis-associated cell population dynamics and molecular signatures in the human and ... https://t.co/f27tKkxQMI
#biorxiv_genomic
0
3
3