The Zhou Lab
@zhou_lab
Followers
2K
Following
277
Media
9
Statuses
88
We are the Zhou Lab @UCLA. We use #cryoEM and #cryoET to study viruses, viral proteins, protein assemblies, and membrane proteins. Student-curated account.
University of California, Los
Joined May 2019
We're so proud to be featured on the cover of @nature! Brilliant work from Dr. Peng Ge and team, uncovering intricacies of the fascinating P. aeruginosa bacteriocin at high resolution using #cryoEM.
This week on the Nature cover: Molecular syringe. High-resolution structures shed light on the workings of an antibacterial nanomachine. Browse the issue here: https://t.co/hX97FaLYX2
2
53
331
Here is the link to the article. https://t.co/X827BSBs6y We are also excited to see the accompanying paper on the doublet structure of trypanosomatid parasites from the labs of Rui Zhang and Alan Brown. https://t.co/ndzOOrEOle
science.org
The movement and pathogenicity of trypanosomatid species, the causative agents of trypanosomiasis and leishmaniasis, are dependent on a flagellum that contains an axoneme of dynein-bound doublet...
0
0
1
Our study on the atomic structure of the T. brucei microtubule, has been published in Science and featured in UCLA News! This work was led by Dr. Xia @xianxia13 in collaboration with colleagues from Dr. Kent Hill's and Dr. James Wohlschlegel's labs. https://t.co/z0eWGQJrVT
2
6
28
Excited to share our latest work: Using cryoET, we unraveled the in situ structure of a methanogen surface layer and its cell envelope. Now published in Science Advances! #teamtomo
science.org
Cryo–electron tomography of the cylindrical archaeal cell resolves the surface layer structure and subunit interactions.
0
6
37
Please check out our just published Nature paper "Native DGC structure rationalizes muscular dystrophy-causing mutations", the excellent teamwork from @ShihengLiu @ tiantiansu and @xianxia13
Thrilled to share our latest work: native cryo-EM structure of the Dystrophin-Glycoprotein Complex (DGC) is now published in Nature! https://t.co/UhiQFq6xbS This discovery sheds new light on the molecular basis of muscular dystrophies and pave the way to novel therapies.
0
3
29
Excited to share our new work! We shed light on the sociology of virus-induced condensates supporting reovirus assembly and replication. Visualized using #CryoET #CryoEM and #FIBmilling
https://t.co/yYjyIipgyj Great collab w/ @xianxia13 @zhou_lab Xiaoyu Liu @gonenlab
nature.com
Nature Communications - Virus-induced cellular condensates are poorly understood. Using cryoET and cryoEM, the authors visualized the 3D “molecular sociology” of host-virus interactions...
0
4
17
Proud to see the spIsoNet is online now! Hope you enjoy using it!
Introducing spIsoNet, a software tool that addresses the challenges of map anisotropy and particle misalignment in cryo-EM caused by the preferred orientation problem. @zhou_lab @yuntaoinnit @hongcheng_fan
https://t.co/vViLnZWwie
0
18
97
We have a fully-funded postdoc position in the @cmholab in #NYC to study how #membraneproteins drive host-pathogen interactions in #malaria parasites using #CryoEM and #CryoET!🦠❄️🔬Email chi-min.ho@columbia.edu with your CV to apply! https://t.co/jZa2WxFLFk
#teamtomo
1
86
156
spIsoNet is available on Biorxiv and GitHub now!
Excited to read this from @yuntaoinnit in the @zhou_lab - saw Hong present it at last year's 3DEM GRC and have been waiting to try it out, look forward to trying spIsoNet on our own data soon! 🤩 https://t.co/OBt9MgDbfu
1
13
47
Beautiful work from Xian @xianxia13 and our amazing collaborators!
Excited to share our latest work in @CellCellPress on how bluetongue virus packages its RNA & assembles its capsid inside host cells! Using #cryoET & #cryoEM, we've visualized this process with 11 structures. Great collab w @gonenlab @zhou_lab 🦠🔬 https://t.co/RvDcVGLyUL
0
2
22
Our preprint of TomoNet is online now!
TomoNet: A streamlined cryoET software pipeline with automatic particle picking on flexible lattices https://t.co/8j2TgsV7RF
#biorxiv_cellbio
1
2
12
Check out our latest preprint work on SARS-CoV-2 Nsp15: https://t.co/v33tg9euo5 We present cryo-EM structures of SARS-CoV-2 Nsp15 bound to its physiological substrate polyU containing RNA, illuminating the basis for immune evasion tactics by coronaviruses!
1
8
63
Our paper is out! We resolved the sheath structure from M. Hungatei cells using cryoET. Thanks to all the coauthors @zhou_lab and Gunsalus lab.#TeamTomo
nature.com
Nature Communications - Cellular cryoET reveals how an amyloid-like protein of the prototypical archaeon, Methanospirillum hungatei, oligomerizes into a ring containing a giant 2700-strand β...
2
19
46
We're also recruiting postdocs! DM or email if you'd like to join us in using integrated structural biology approaches combining #CryoEM and #insitu #CryoET to study #membraneproteins and molecular machines in #malaria parasites! Check it out👇👉 https://t.co/jZa2WxFdPM
#teamtomo
4
90
246
Congrats!
Check out our latest paper on HIV Vif-APOBEC3H complex structure by cryo-EM: https://t.co/gu1rBuV8iv This has been a collaborative project between @zhou_lab and Xiaojiang Chen lab at USC.
0
0
4
Check out the recent cryoET work with our beloved colleagues in Hill lab on the flagella structure of T. Brucei.
nature.com
Nature Communications - Microtubule inner proteins (MIPs) contribute to species-specific motility characteristics but are largely unstudied. Here, the authors combine functional, structural and...
2
5
16
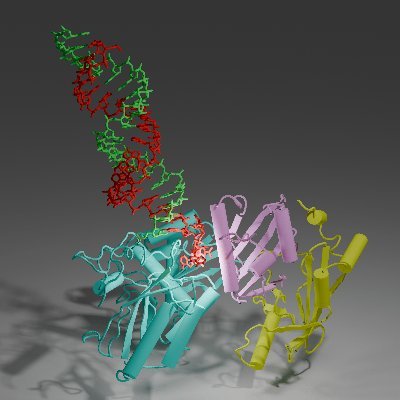
Check out our cryoEM result of mRNA editosome in Trypanosoma brucei! Awesome collaboration with Aphasizhev lab from Boston U. Many congrats to Bruce @ShihengLiu and other coauthors for another wonderful piece of work!
science.org
Structural analysis of the RNA editosome reveals mechanisms of guide RNA recognition and messenger RNA engagement.
4
17
63
Congrats to Jason, Jane and Yuntao @yuntaoinnit
The group of Hong Zhou @MicrobioUcla & their collaborators determined the cryo-EM structure of filamentous 3-methylcrotonyl-CoA carboxylase from Leishmania tarentolae and discuss its mechanism @zhou_lab
https://t.co/gdL7WepqaO
0
2
8
Long-lasting enigma in HIV study: how HIV antagonizes the host antiviral systems and evades the immunity? We are addressing the question by cryo-EM structure of Vif-APOBEC3G complex, which emerged as an unusual ribonucleoprotein complex: https://t.co/JiO1lWMMxn
0
4
20
Atomic model of vesicular stomatitis virus and mechanism of assembly @zhou_lab
#virology #CryoEM #structures
https://t.co/jjWNxBuiYM
0
10
32