Jonas Demeulemeester
@zeunas
Followers
547
Following
1K
Media
6
Statuses
559
Group Leader at the VIB – KU Leuven Center for Cancer Biology @vib_ccb and @Onco_KULeuven interested in tumour evolution, single-molecule multi-omics, and AI
Leuven, Belgium
Joined November 2012
Big, beautiful trees!! SMART-PTA for whole-genome+transcriptome on thousand of single cells from the normal human esophagus 🤯 Massively scaling up the power of scWGS to build deep phylogenies and chart somatic evolution from birth throughout life. https://t.co/pciw5yME0x
14
74
245
Our latest work is out in Nature today. In this paper, we introduce an improved version of NanoSeq, a duplex sequencing protocol with <5 errors per billion bp in single DNA molecules, and use it to study the oral somatic mutation landscape in >1000 people.
nature.com
Nature - A new version of nanorate DNA sequencing, with an error rate lower than five errors per billion base pairs and compatible with whole-exome and targeted capture, enables...
6
60
242
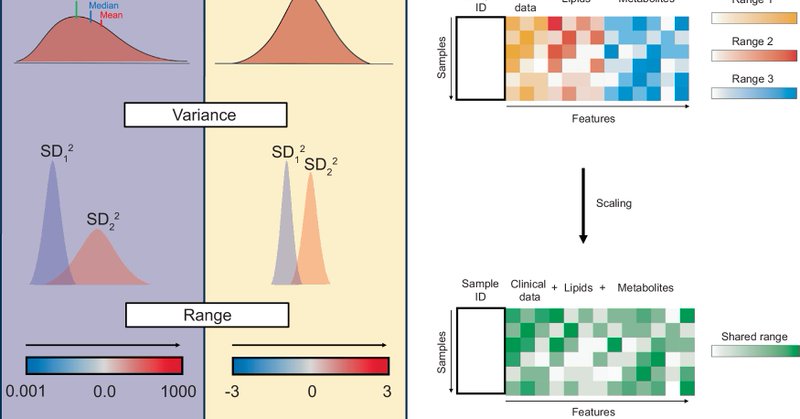
Our new article has just been published in Nature Communications: “Best practices and tools in R and Python for statistical processing and visualization of lipidomics and metabolomics data” https://t.co/PA4fl5ULJJ
#Lipidomics #Metabolomics #OpenScience #RStats #Python
nature.com
Nature Communications - Mass spectrometry-based lipidomics and metabolomics generate large, complex datasets requiring effective analysis. Here, authors review key statistical and visualization...
0
9
13
1/ Happy to share that our first preprint from the Demeulmeester lab (@zeunas) is out now! We present our new multi-modal single-cell long-read method: SPLONGGET (Single-cell Profiling of LONG-read Genome, Epigenome, and Transcriptome) https://t.co/fi7iroCE0o
1
2
5
Proud to share our first TRACERx cancer study integrating epigenomics + genomics in NSCLC as co-drivers of tumor evolution. Led by not me but by the wonderful @NnennayaKanu, @F_gimenoval, @ccastignani6, @VanLooLab
2
80
262
I’m excited to share the first manuscript out of my PhD work with @NAltemose, in which we describe FiberFold: a deep learning tool that predicts cell-type-specific and haplotype-specific 3D genome organization from a single experiment! https://t.co/FwJldYnCGe 1/
biorxiv.org
The three-dimensional (3D) architecture of the genome plays a crucial role in gene regulation and various human diseases. Short-read sequencing methods for measuring 3D genome organization are...
1
11
32
Thrilled to share that my 2nd first-author paper from my PhD is now published in @NAR_Open! After >9 years of work and an amazing collaboration with @k_theun, this project holds a special place in my heart (1/9) #SingleCell #MultiOmics
https://t.co/OpolVh1ZPz
academic.oup.com
Abstract. Single-cell multi-omics methods enable the study of cell state diversity, which is largely determined by the interplay of the genome, epigenome,
2
1
3
We zijn er klaar voor. Welkom op campus Gasthuisberg,ON4,7de verdieping voor 600 jaar KU Leuven en onze bijdrage aan kankeronderzoek! @vib_ccb @LKILeuven @ChristineDesme2 @cancerinpregnancy.org
1
1
2
I am looking for a postdoc to join my new team in Melbourne, Australia at WEHI to work on lung stem cells and disease, including lung cancers! International candidates+ those just finishing up PhDs welcomed Please apply below: https://t.co/VUVq7XwoMf
4
87
177
Spread the word! Registration for the "Mutations in Time and Space" conference is open. The meeting is all about the origins, patterns, and consequences of mutations across cells, individuals, populations, species. Abstract submission deadline: Jan 17th https://t.co/KMEHdXnLMQ...
1
22
51
📢Very excited to share this paper with you! The oestrous cycle stage affects mammary tumour sensitivity to chemotherapy published @Nature . Congratulations to Laura, @ColindaScheele and all the authors for this amazing story. https://t.co/UvdwYsJMK8
nature.com
Nature - In three mouse models of breast cancer, we show reduced responses to neoadjuvant chemotherapy when treatment is initiated during the dioestrus stage, when compared with initiation during...
8
43
139
🚨So proud to present the first study from our @CCG_UCL group together with @CharlesSwanton @NnennayaKanu and led by @ojlucas with key collaboration @Ward__Sophie and fundamental support @Rija_Zaidi_ @abi_bunkum & many others Out in @NatureGenet 👇 https://t.co/cd7YkBBDxo 🧵👇
6
39
162
The latest hifiasm can directly assemble standard @nanopore simplex R10 reads, without HERRO correction or other preprocessing, to phased contigs of contiguity comparable to HiFi assembly. Like before, you can further add ultra-long, Hi-C or trio data for better assembly.
Exciting news! The latest hifiasm release from @ChengChhy and @lh3lh3 adds beta support for @nanopore simplex R10 reads. Initial results look very promising. 🚀 Check it out:
https://t.co/Ktz0102BHL"
2
58
184
Deaminase-assisted Fiber-seq (DAF-seq) has arrived! DAF-seq is an amplification-compatible version of #Fiberseq that resolves genetic and chromatin architectures with single-nucleotide, single-molecule, single-haplotype, and single-cell precision.
biorxiv.org
Gene regulation is mediated by the co-occupancy of numerous proteins along individual chromatin fibers. However, our tools for deeply profiling how proteins co-occupy individual fibers, especially at...
1
30
93
Hot off the press!! 🔥 Check out our work on the origins and impact of ecDNA. This was a team effort involving multiple collaborators as one of a three part series on ecDNA in @Nature
ecDNA are rogue genetic material that amplify genes, contribute to treatment resistance and drive genomic heterogeneity. We report on the Origins and impact of extrachromosomal DNA today in @Nature
5
12
69
In just two weeks, we’re gathering an exceptional community of researchers and innovators at the intersection of AI and life sciences in Mechelen for our inaugural symposium. We are grateful to our sponsors: ✨ ICOSA ✨ Johnson & Johnson Innovative Medicine See you soon!
0
1
2
Very happy and excited to announce that I'll be starting my own research group at @emblebi! The group will focus on lineage tracing, somatic evolution and the origins of cancer. Interested in doing a postdoc in the group or know someone who is? Please reach out!
41
31
262
Last week we celebrated moving into our brand-new lab space in ON6! 🥳 There are still some boxes to be unpacked, but we look forward to doing some amazing science here. PS: We're still hiring... https://t.co/5tPNhRYXOS
0
3
13
🦍Postdoc in Somatic Evolutionary Genomics🦍 Join my team @GeneticsCam on a pioneering project mapping somatic mutations across primates! 🌍Collaborate with experts @UCBerkeley & @sangerinstitute. 🔍Experience in genomics? Apply by Nov 16! Details: https://t.co/1pDO1tzzcP
0
74
126
Which method works better for single-cell Whole Genome Amplification? By comparing the 3 methods, we found that the answer depends on what matters most in each downstream application. A big thanks to @proukakis, @sedlazeck, @zeunas, @ASAP_Research, @QSBrainBank , @UCLIoN
longest amplicobs, useful for long read sequencing (watch this space!). Great lab work by @Kalef_Ezra and bionformatic guidance by @sedlazeck @zeunas, funded by @ASAP_Research , tissue from @QSBrainBank at @UCLIoN
https://t.co/tbAzrNL63t
0
2
5