Peter Van Loo
@VanLooLab
Followers
2K
Following
11
Media
1
Statuses
50
Professor in cancer genomics and tumor evolution at MD Anderson Cancer Center Visiting Group Leader at the Francis Crick Institute
Houston
Joined March 2019
🚀 Excited to share our latest research! We developed GRITIC, a novel method for timing genomic gains using bulk WGS data. Our findings reveal non-parsimonious evolution and increased chromosomal instability post-genome doubling in tumors. #cancergenomics.
0
27
93
RT @NatureCancer: NEW REVIEW ARTICLE @NatureCancer – “Aneuploidy and complex genomic rearrangements in cancer evolution” by @VanLooLab #Tob….
0
39
0
Exciting find in our #OralCancer study! 🧬 Brush biopsies prove promising, reflecting 90% of somatic changes vs. tissue biopsies. 🩸 Highlighting their potential in understanding tumor evolution. #CancerEvolution
0
21
71
RT @mvicaracal: Today, @nature and @NatureMedicine are proud to publish amazing new insights into lung cancer evolution from the #TRACERx c….
0
74
0
RT @kandymarsy: World class TRIUMPH mentors are hiring post-doctoral fellows @MDAndersonNews: @JenWargoMD @swatowic @rkalluriMDPhD @Tanigu….
0
7
0
RT @rfschwarz: How can we track cancer evolution using somatic copy-number alterations while considering whole-genome doubling events? .Wit….
0
27
0
RT @TheCrick: Registration is now open for our Somatic Evolution and Tumour Microenvironment (SETM) symposium, where discussions and talks….
0
5
0
@DrewsRM et al. deconstructed chromosomal instability in 6,335 cancers to discover 10 pan-cancer and 7 cancer-type-enriched signatures. CIN signatures associated with known aetiologies - replication stress, mis-segregation and impaired recombination: (3/4).
1
5
29
@cd_steele et al. investigated allele-specific copy number signatures across human cancers. Copy number profiles from 9,873 cancers in TCGA were decomposed to reveal 21 signatures, many with a clear mechanistic origin: (2/4).
1
5
34
For years, we’ve looked for recurrent copy number changes in cancer but lacked a pan-cancer view of the processes that give rise to these events. We've collaborated on two @Nature studies into the mechanisms underlying aneuploidy and chromosomal instability (1/4).
2
72
298
Total mRNA expression in tumor cells is an important, but so far elusive, biomarker. This elegant piece of team science establishes the mathematical relationships between copy number, purity and gene expression in tumor cells. Proud to have been part of this great team!.
Very excited to share our paper in Nature Biotechnology today! Huge amount of work by @ShaolongCao @sxj307 @JenniferWangMD et al! It is amazing to work with many experts across cancers. Every cancer tells its own story and our metric TmS can quantify it!
0
8
76
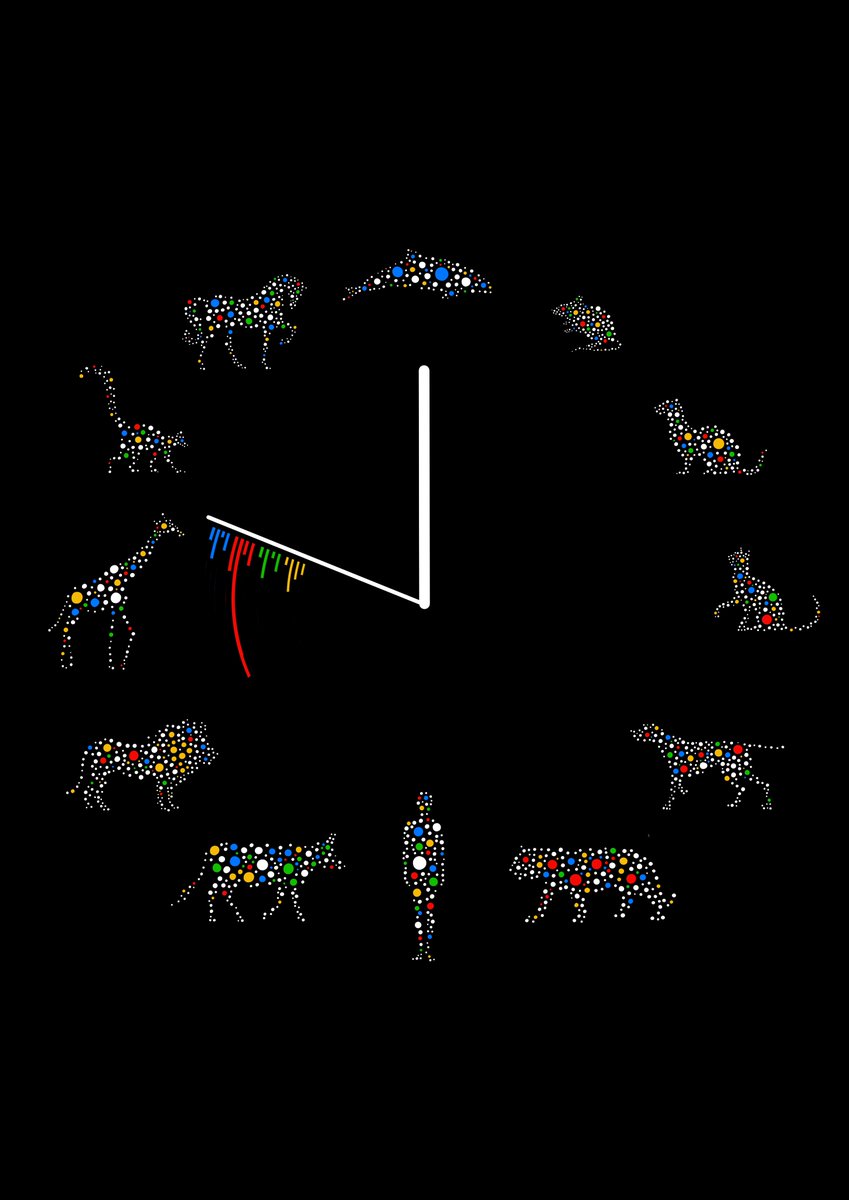
Fantastic work linking somatic mutation rates with lifespan across mammalian species, allowing us to make sense of the lack of relationship between body size and cancer incidence.
Incredibly excited to share our paper ‘Somatic mutation rates scale with lifespan across mammals’ now published @nature. An illustrated & updated tweetorial… [1/24]
0
0
11
We’re looking for #postdocs interested in #cancergenomics to join our lab at @MDAndersonNews – An exciting opportunity to lead large-scale studies of #tumorevolution:
1
33
73
RT @vib_ccb: We are delighted to announce Dr. Jonas Demeulemeester as a new group leader in the @VIBLifeSciences Center for Cancer Biology….
0
11
0
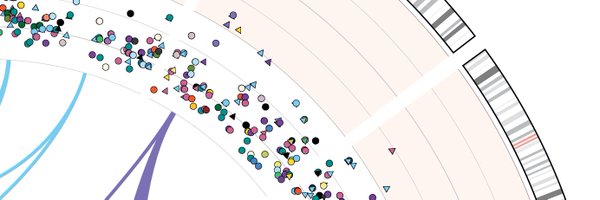
Excited to share our new paper in @NatureGenetics! Can a mutation really hit the same place twice? We found biallelic mutations across 2,658 cancer genomes that contradict the famous infinite sites assumption and highlight the processes behind them:
3
96
373