Yotam Drier
@yotam_d
Followers
346
Following
244
Media
1
Statuses
395
Genomics, Epigenomics and Topology of Cancer • Assistant Professor, Faculty of Medicine, Hebrew University of Jerusalem • https://t.co/wi6QPVtVg2
Jerusalem
Joined September 2009
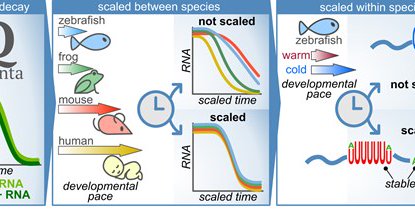
RT @MichalRabani: How embryos stay “on time” as they grow?.Happy to share our recent publication: Quantitative modeling of mRNA degradation….
academic.oup.com
Abstract. As embryos transition from maternal to zygotic control, precise clearance of pre-loaded maternal mRNAs is essential for initiating new zygotic ge
0
14
0
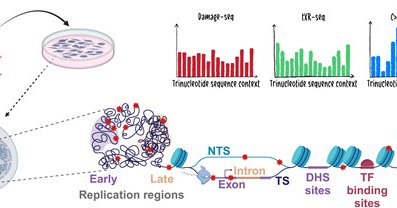
RT @sheeradar: New paper alert! The epigenetic landscape shapes smoking-induced mutagenesis by modulating DNA damage susceptibility and DNA….
academic.oup.com
Abstract. Lung cancer sequencing efforts have uncovered mutational signatures that are attributed to exposure to the cigarette smoke carcinogen benzo[a]pyr
0
9
0
RT @AkisPapantonis: The @RoukosVassilis lab led a new and detailed study of Topoisomerase II contribution to #3Dgenome folding in human cel….
0
21
0
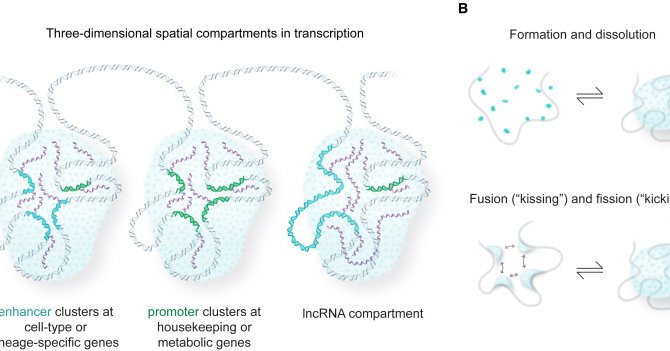
RT @jehenninger: Check out our review on an RNA-centric view of transcription and genome organization in an #RNA-focused issue of @Molecula….
cell.com
RNA molecules are involved in diverse regulatory mechanisms that contribute to gene activation, gene repression, and genome structure. Henninger and Young discuss how the more traditional DNA-prote...
0
23
0
RT @jane_skok: Thrilled to share our new preprint, 'A genome wide code to define cell-type specific CTCF binding and chromatin organization….
biorxiv.org
CTCF-mediated chromatin folding plays a key role in gene regulation, however the mechanisms underlying cell type-specific control are not fully elucidated. Comprehensive analyses reveal that CTCF...
0
30
0
RT @peterwlaird: Excited with to share our new deep single-cell whole genome bisulfite sequencing technique with @shenhui1986, along with s….
0
13
0
Check out the newest version of our paper, now published in PNAS: A good way to start the new Hebrew year. Congratulations to all authors. More details in the thread below.
A new manuscript in these difficult times: We profiled the cis-regulatory landscape of lung neuroendocrine tumors in collaboration with @simonag56739514. We uncovered a subtype resembling hepatic differentiation due to HNF1A and HNF4A activation, . 1/3.
3
1
17
RT @ElenaPugachev12: CTCF binding landscape is established by the epigenetic status of the nucleosome, well-positioned relative to CTCF mot….
biorxiv.org
CTCF binding sites serve as anchors for the 3D chromatin architecture in vertebrates. The functionality of these anchors is influenced by the residence time of CTCF on chromatin, which is determined...
0
16
0
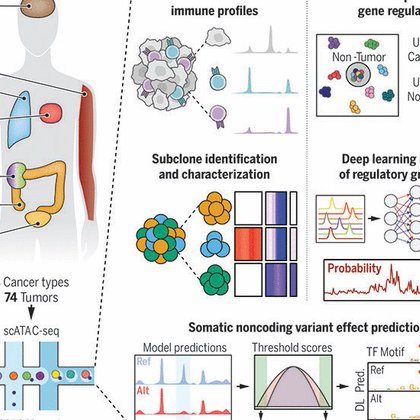
RT @WJGreenleaf: Our single-cell, pan-cancer analysis of open chromatin in human cancers, done in collaboration with TCGA and the Chang and….
science.org
To identify cancer-associated gene regulatory changes, we generated single-cell chromatin accessibility landscapes across eight tumor types as part of The Cancer Genome Atlas. Tumor chromatin...
0
81
0
Great opportunity for postdocs! If you are interested in epigenetic and topologic dysregulation in disease consider joining our lab, see for more details. We will help you apply to the Azrieli Fellowship :).
Attention prospective postdocs!. The Azrieli International Postdoctoral Fellowship for Research at Israeli Universities in STEM, Humanities, and Social Sciences is now open for applications!. You can apply today for the 2025–26 cohort. The Azrieli International Postdoctoral
0
0
1
RT @wamoyal: Great new paper using #3D #MERFISH to generate 3D genome cancer atlases of murine lung and pancreatic adenocarcinoma. Tracing….
biorxiv.org
Although three-dimensional (3D) genome structures are altered in cancer cells, little is known about how these changes evolve and diversify during cancer progression. Leveraging genome-wide chromatin...
0
2
0
RT @fabian_theis: Want to learn how transformers and large-language models can help to organize&understand single cell profiles?.Led by @ar….
0
90
0
RT @Hodges_Lab: In this new preprint, we do a deep dive into the temporal relationship between DNA #methylation, #chromatin and #enhancers.….
biorxiv.org
Epigenetic mechanisms govern the transcriptional activity of lineage-specifying enhancers; but recent work challenges the dogma that joint chromatin accessibility and DNA demethylation are prerequi...
0
23
0
RT @Noam_Kaplan: The radial positioning of genomic loci, i.e. how far they are from the nuclear lamina has been associated with genome stru….
0
17
0
Thank you @LautenbergI ! and many thanks to all the students in my lab whose work made this possible.
Congratulations to our esteemed member, Dr. Yotam Drier, for winning the prestigious Young Researcher Award at the 2024 Faculty of Medicine Awards! 👏 Your hard work and dedication are truly inspiring.
1
1
22
RT @anshulkundaje: Really awesome paper from @gagneurlab on a new, clever interpretation approach showing that local DNA language models (t….
biorxiv.org
Deciphering how nucleotides in genomes encode regulatory instructions and molecular machines is a long-standing goal in biology. DNA language models (LMs) implicitly capture functional elements and...
0
22
0
RT @bvansteensellab: We generated a collection of new and optimised (and multiplexable) TF reporters for human and mouse cells: https://t.c….
biorxiv.org
In any given cell type, dozens of transcription factors (TFs) act in concert to control the activity of the genome by binding to specific DNA sequences in regulatory elements. Despite their conside...
0
12
0
RT @bvansteensellab: The first preprint of our PERICODE consortium: MPRA-trained deep learning provides insight into the regulatory logic….
0
16
0