Tschaharganeh Lab
@tschaharganeh
Followers
323
Following
228
Media
44
Statuses
186
The Tschaharganeh lab @uniklinik_hd and @DKFZ investigates causes and cures of cancer using new methods to interrogate genetic information
Heidelberg, Germany
Joined September 2018
0/ Preprint alert! 🚨 One step closer to solving the genotype ➡ phenotype puzzle in cancer! Led by @BreinigMarco, @LomakinAI, and @HeidariElyas, our novel approach, #CHOCOLAT_G2P 🍫, deciphers the impact of combinatorial alterations on tumor phenotypes and their environment.
Integrated combinatorial functional genomics and spatial transcriptomics of tumors decodes genotype to phenotype relationships https://t.co/J5lwI4RpTp
#bioRxiv
1
21
42
We are happy to share our latest paper @natBME spearheaded by @BreinigMarco. Together with @MoritzGerstung we hack commercial #spatialtranscriptomics @10xGenomics to decode #phenotypes from #genotypes in #cancer: https://t.co/JTXzMkDiCW
@DKFZ @uniklinik_hd
nature.com
Nature Biomedical Engineering - A method integrates perturbation mapping with 10X Visium spatial transcriptomics to map tumour genetic complexity and heterogeneity.
0
3
9
Thrilled to receive an #ERCCoG grant from @ERC_Research! Huge thanks to my amazing lab members—past and present—my family, and the incredibly supportive environment @DKFZ @KiTZ_HD. Excited to advance research on pediatric sarcomas. Stay tuned, and congrats to all awardees!
📢 Results of the 2024 ERC Consolidator Grant competition are out. €678 million for 328 researchers. Who won funding? What will they investigate? Where will they conduct their research? #ERCCoG news 👉 https://t.co/BB63zeBC2x 🇪🇺 @HorizonEU
19
7
64
Double ERC success for DKFZ researchers: Ana Banito (@KiTZ_HD / DKFZ) and Aurélie Ernst (DKFZ) receive the prestigious ERC Consolidator Grant. Congratulations! 👏 #ERCCoG @ERC_Research ➡️ https://t.co/G9hmaVToqc
4
6
33
And here we present our newest #phd!! Sonia defended her work on chromosomal amplifications @DKFZ @uniklinik_hd!! Great defense and we are looking forward to an important paper using #Genetics and #immunotherapy for #therapy of #Cancer. More to come soon!! @DKFZImmunology @NCT_HD
3
2
18
2/📢 @skrieg1990 first author paper is out! Congrats 🎉 BIG THANKS TO ANYONE INVOLVED!🙏 @dkfzimmunology @DKFZ @uniklinik_hd Great collab with @maxbillmann! Thanks to @GenomeMedicine and reviewers for helping us improve our work!
1
0
5
1/🥳Paper alert! https://t.co/wxKCaodlxD In(Fe)rring synthetic lethality with chromosome 8p deletions - learn how we 🎯 paralog synthetic lethality 🧬☠️ involving mitochondrial iron transporters MFRN1 and MFRN2 to attack cancer cells with specific somatic copy number alterations.
1
3
7
Krieg and coll. use genetic and computational approaches to show that Mitoferrin2 can be targeted as synthetic lethality in cancers bearing chromosome 8p deletions. @maxbillmann @BreinigMarco @tschaharganeh Full paper here: https://t.co/UEL1PEi6j1
genomemedicine.biomedcentral.com
Background Somatic copy number alterations are a hallmark of cancer that offer unique opportunities for therapeutic exploitation. Here, we focused on the identification of specific vulnerabilities...
1
8
10
Today we celebrate Aga’s #phd defense @DKFZ @UniHeidelberg @uniklinik_hd. She did an amazing job with her project on #livercancer #cholangiocarcinoma. Glad you will stay for longer in the lab. #phdone #Good_Performance
2
3
15
This is a really cool colab of ours investigating spatial cancer biology in vivo with with a massively parallel genetic perturbation approach that runs on the back of 10x Visium. Check out the video for a quick summary and preprint for details.
0/ Preprint alert! 🚨 One step closer to solving the genotype ➡ phenotype puzzle in cancer! Led by @BreinigMarco, @LomakinAI, and @HeidariElyas, our novel approach, #CHOCOLAT_G2P 🍫, deciphers the impact of combinatorial alterations on tumor phenotypes and their environment.
1
1
10
12/ Absolutely thrilled for this remarkable collaborative effort among labs. A heartfelt thank to @GerstungLab @TschaharganehLab @StatGenomics @sahm_lab and everyone involved for their invaluable contributions and support throughout this 🍫 journey. @uniklinik_hd , @DKFZ, @EMBL
0
0
3
11/ We share our findings through an interactive web browser: https://t.co/XbjKDKDtPp. For a deeper dive, read our preprint. Check out our code on GitHub: https://t.co/II7mFisaUx. Raw data available at https://t.co/RsK39DJ7Xe. Happy exploring!
1
0
1
10/ Our method linked genotypic variations to diverse tumor phenotypes, revealing intriguing connections. Notably, we found strong associations between cholangiocarcinoma and VEGFA, alongside negative links with mtCtnnb1, linking genetic alterations and tumor phenotypes.
1
0
0
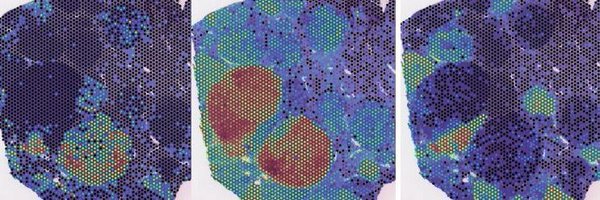
9/ #CHOCOLAT_G2P revealed the diverse landscape of tumors and their microenvironments, distinguishing liver tumor subtypes and zonation patterns. It highlighted the spatial patterns of immune cells and cancer-associated fibroblasts (CAFs).
1
0
1
8/ Our genotyping approach uncovered codependency patterns and mutual exclusivity among certain alterations. Notably, Myc and mtCtnnb1 combinations were predominant. Surprisingly, despite its recognized significance in human liver cancer, VEGFA did not play a significant role.
1
0
1
7/ Consequently, we computed the probability for all 256 possible genotypes defined by eight perturbations. This transformation process converted spatial barcode signals into genotypically-defined clonal maps.
1
0
0
6/ Across the 324 identified nodules, our Bayesian model effectively calculated probabilities for the existence of each of the eight perturbations.
1
0
0
5/ #CHOCOLAT_G2P experiments yielded an astonishing 324 tumor nodules in just ~2.5 cm2 sectioned tissue, enabling us to study a wide range of co-existing tumor genotypes and phenotypes.
1
0
0
4/ To conveniently leverage existing technology in a “plug-and-play” fashion, we repurpose 10X Visium RTL-probes designed to detect chemosensory receptor transcripts. We craft 50nt molecular barcodes from olfactory, taste, and vomeronasal receptor capture sequences.
1
0
1
3/ To facilitate orthogonal spatial transcriptomics analysis, we utilised molecular barcoding aligned with RNA-templated ligation (RTL) probe capture, integrated within the commercial 10X Visium for FFPE kit. @10xGenomics
1
0
0
2/ We modify an established autochthonous liver cancer mouse model to introduce higher-order combinatorial perturbations. Utilising molecular barcoding technology, we subsequently identify and analyse the resulting alterations.
1
0
0