Stegle Lab
@StatGenomics
Followers
3K
Following
27
Media
1
Statuses
62
We're moving to Bluesky - come join us: https://t.co/nHQ8M13K8Z
Heidelberg, Germany
Joined September 2013
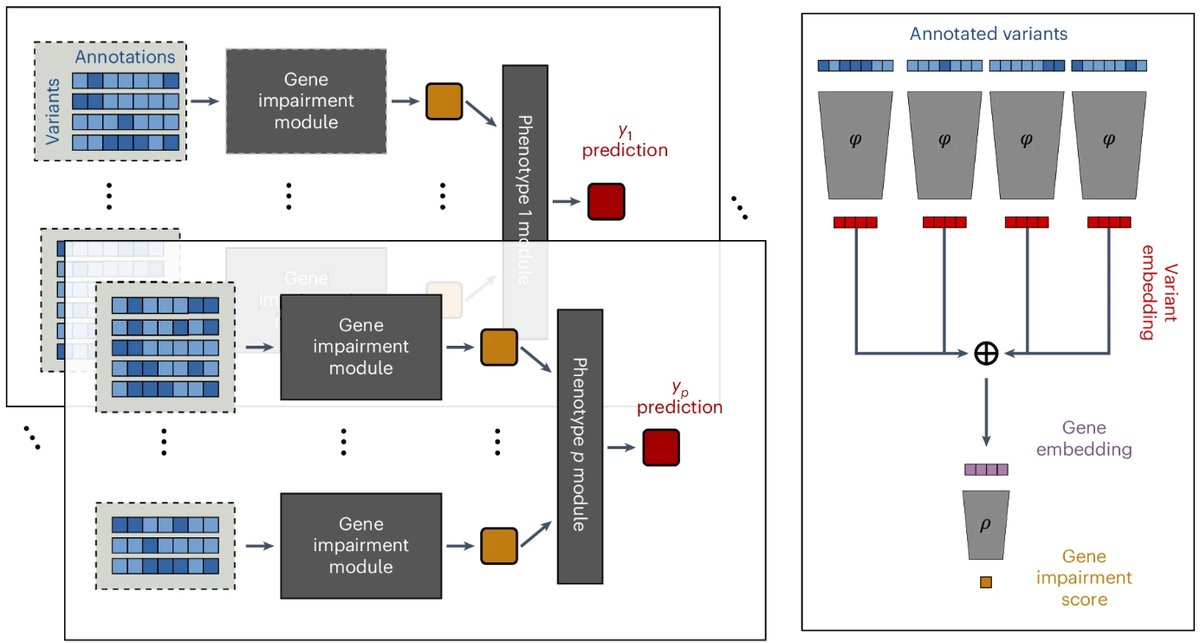
Our paper on using machine learning to predict the impact of rare genetic variants is out! #DeepRVAT utilizes a fully data-driven approach to analyze annotated variants. @DKFZ press release: Publication in @NatureGenet:
0
8
34
We are looking for a postdoctoral researcher! .Join our translational project aimed at exploring the processes that alter the epigenetic landscape of colorectal cancer cells and drive their metastasis. Become part of our group at @EMBL .Apply here:
0
9
16
Amazing work from our #DeepRVAT team! Using AI to predict the effects of rare variants, pushing the boundaries of personalised medicine. 🧬🔍.Check out the thread and explore their exciting results yourself!.
🧵 1/9 Delighted to announce that #DeepRVAT is out today in Nature Genetics! Here's a recap of our tweetorial from the preprint, with updates for some exciting new results. It's been a joy to work with @Holtkamp_Eva, as well as @gagneurlab and @OliverStegle.
0
0
5
RT @DKFZ: Apply now for the DKFZ #Postdoc|toral Fellowships at the German #Cancer Research Center, covering all areas of #cancerresearch, i….
0
11
0
Do you want to pursue a PhD at the intersection of AI and the life sciences? 🤖🧬. We are recruiting through the @hidss4health program! Join an exciting project on multi-omics factorization models for personalized medicine with @JunyanLu1118. Apply here:
hidss4health.de
0
2
6
Congratulation, Eva!.
Feeling extremely honored to receive the #eshg2024 Early Career Poster Award! This work wouldn’t be possible without everybody involved in DeepRVAT, esp. @brianfclarke @gagneurlab @StatGenomics
0
0
6
#CHOCOLAT_G2P 🍫🐭 is available as a preprint! Our collaborative project on tumor spatial genomics, showcases a new technology enabling induction and spatial genotyping/phenotyping of hundreds of tumors in mouse liver. Check out the thread for the nuts and bolts of 🍫-G2P!.
0/ Preprint alert! 🚨 .One step closer to solving the genotype ➡ phenotype puzzle in cancer! Led by @BreinigMarco, @LomakinAI, and @HeidariElyas, our novel approach, #CHOCOLAT_G2P 🍫, deciphers the impact of combinatorial alterations on tumor phenotypes and their environment.
1
1
9
🌟 Be part of our team!.Join our lab for a postdoc in computational biology, focused on multi-omics profiling in time and space. Apply now:
0
10
14
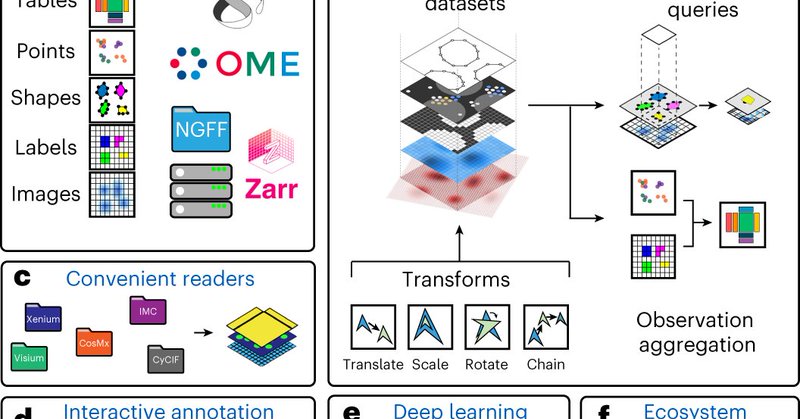
One framework to rule them all: SpatialData 🔳 . Excited to see our efforts in unifying and integrating multimodal spatial omics data highlighted by @embl. Phenomenal teamwork with @fabian_theis and @notjustmoore and more! 💪.Published in @naturemethods: .
nature.com
Nature Methods - SpatialData is a user-friendly computational framework for exploring, analyzing, annotating, aligning and storing spatial omics data that can seamlessly handle large multimodal...
SpatialData is a tool developed by EMBL scientists in cooperation with multiple research institutions to unify & integrate data from different omics technologies. It provides holistic insights into health & disease and is publicly accessible.
1
7
25
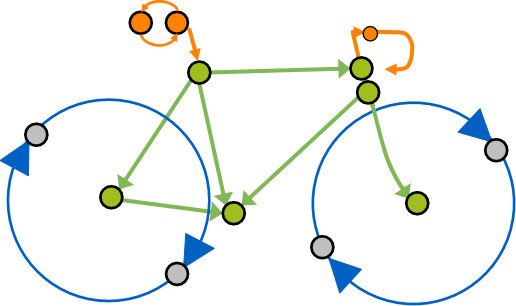
Two wheels, one method to infer causality in biological networks: Bicycle! 🚲.Thrilled to share our team's breakthrough. Dive into the details in the thread below 👇.
🧵1/10 We’re thrilled to share our latest paper, Bicycle, a novel approach for inferring cyclic causal relationships from observational and interventional data! This work addresses a critical gap in understanding complex biological networks.
0
2
12
We are seeking a postdoctoral researcher at @EMBL!! 🦸♀️.Are you passionate about applying machine learning to unravel the causal mechanisms of gene regulation? .Apply now:
0
3
6
Two papers up in @NatGenet on the same day!.1) Population-scale single-cell RNA-seq profiling across neuron differentiation 2) Identification of regulatory variants in pluripotent cells using population-scale transcriptomics
3
32
133
New lab paper up. Work by @BrittaVelten. We married up GPs with sparse factor analysis, which turns out to be a great fit for dealing with spatio-temporal data.
Are you working with longitudinal/temporal/spatial omics data? Have a look at our new preprint: We present MEFISTO a method for the functional integration of spatial and temporal multi-omics data. Joint work with @OliverStegle @RArgelaguet.
0
0
2
RT @OliverStegle: New Paper up! Great multi-lab team effort with @gagneurlab @anshulkundaje to establish a repository of ML models in genom….
0
96
0
New lab paper is up. StructLMM is a method to jointly test for genotype-environment interactions using tens or hundreds of exposures. Yields new GxE signals for BMI and gene expression traits. By @rachelmoo and @fpcasale with Barroso lab at Sanger.
0
19
36
New lab paper together with @sysbiomed. Great collaboration, providing new cues on the germline component of inter individual variation in drug response.
Menden @fpcasale et al. The germline genetic component of drug sensitivity in cancer cell lines - joint work w @StatGenomics
0
1
9
RT @OliverStegle: Paper on interpretable and sparse factor analysis for scRNA-seq is up. Generalises scLVM; software for R & python! https:….
genomebiology.biomedcentral.com
Single-cell RNA-sequencing (scRNA-seq) allows studying heterogeneity in gene expression in large cell populations. Such heterogeneity can arise due to technical or biological factors, making decomp...
0
39
0