Stephanie Lo
@stephlo_lo
Followers
1K
Following
2K
Media
56
Statuses
1K
Associate Professor @UniofBath and Senior Staff Scientist for Global Pneumococcal Sequencing (GPS) project @pneumowatch @sangerinstitute
Cambridge, England
Joined November 2015
Grateful of making this video https://t.co/HaVnOTUBSE to explain my research #BetterVaccineDesign #SaveLives #ReduceAMR
@UniofBath @MilnerCentre
We speak to Dr Stephanie Lo @stephlo_lo from the Department of Life Sciences, about her life-changing research into vaccine implementation and antimicrobial usage. https://t.co/0UwsVSovgQ
@UniofBathSci
0
2
36
Microbial Genomics is looking to appoint a Deputy Editor-in-Chief, who will help develop the editorial strategy and new content for the journal. Applications close on 1 November, more information about the role can be found here:
microbiologysociety.org
0
8
15
It’s such a pleasure to see that #GPS_project 🩻funded by @gatesfoundation has been featured in this AMR article by @sangerinstitute , showing our global efforts on evaluating AMR after vaccine 💉 rollout. https://t.co/VaF5DoPptW
sangerinstitute.blog
Our scientists and collaborators are working together to tackle one of the biggest public health threats: antimicrobial resistance. We are using cutting-edge genomics to hunt down drug-resistant...
1
1
2
🦠🧪🧬 Excited to share that our GPS Pipeline, the portable and scalable genomic pipeline for Streptococcus pneumoniae surveillance, is now published in @NatureComms 🔗 https://t.co/zMpJWPTMEl
#GPS_project
nature.com
Nature Communications - The GPS Pipeline enables accessible and scalable genomic surveillance of Streptococcus pneumoniae. It performs quality control and in silico typing of sequencing reads with...
1
8
8
[New pre-print🚨] When a single genotype (pbp) confer resistance to multiple antibiotics (penicillin and cephalosporins), how do we make sense of genetic changes and their impact on AMR? A new concept - AMR cartography is invented by @Quackscience
https://t.co/8UWQNdHgUr
1
2
2
[🚨 New paper] The first nationwide, longitudinal WGS analysis of Streptococcus pneumoniae in Taiwan (2006–2022). 1,343 isolates from 27 hospitals, we track serotype, AMR, and genomic lineage before and after PCV13. #GPS_project #PCV13_impact #Taiwan
https://t.co/PcRsmAmkov
microbiologyresearch.org
1
5
6
New paper out in Pneumonia. More data from the carriage study in Southampton, UK looking at pneumococci in pre-PCV13 era versus early- and late-PCV13 periods (I also snuck in some bits on other bacteria ... just for a bit of variety) https://t.co/prjdIDU8pc
pneumonia.biomedcentral.com
1
5
11
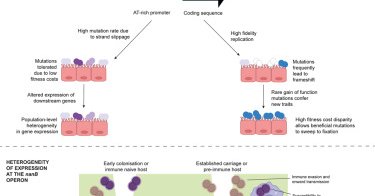
Promoter variation in Streptococcus: S. pneumoniae promoters contain repetitive nucleotides, driving strand slippage & promoter mutations. This drives population-level heterogeneity in gene expression, offering evolutionary strategy to maximize fitness https://t.co/qdojFlpogn
cell.com
Barton et al. identify Streptococcus pneumoniae promoters containing repetitive nucleotide sequences that promote DNA strand slippage and accumulation of promoter sequence mutations. This drives...
0
7
19
A great collaboration with Dr Dan Neill and his team in @dundeeuni and others in @ucl @StJudeResearch @unibern @uniofliverpool @UCT_news @uwanews @UniofBath @MilnerCentre @sangerinstitute @pneumowatch
0
1
1
However, this promoter variant does not thrive on its own and is usually co-occurence of the wild-type promoter without "A", showing heterogeneous gene expression during single infections provides population benefits
1
1
1
This tiny mutation—found not only mouse colonization models but also clinical isolates from the #GPS_project—enhance S. pneumoniae colonization reducing mucus trapping.
1
0
0
[New paper alert🚨] What happens when pneumococci 'slip' and 'add' just one extra A in a promoter spacer of a sugar scavenging gene? 🧬 https://t.co/Sy0fqIPm7S
cell.com
Barton et al. identify Streptococcus pneumoniae promoters containing repetitive nucleotide sequences that promote DNA strand slippage and accumulation of promoter sequence mutations. This drives...
1
5
12
Our review on within-host bacterial evolution is now out @NatureMicrobiol With Gerry Tonkin-Hill, @Josie_M_B @katrina_lythgoe and Stephen Bentley https://t.co/y0GN5bH7Pi
nature.com
Nature Microbiology - In this Review, Tonkin-Hill et al. discuss the processes driving bacterial evolution and emergence of pathogenesis within hosts, the importance of understanding within-host...
0
13
29
Our team is hiring an ambitious senior bioinformatician to look at bacterial capsules . This position will closely collaborate with @emblebi
@StephenBentley5 @johnlees6 @stephlo_lo Closing date: 20 July 2025 https://t.co/pujNp7xiKg
0
10
11
The Rosch Lab is hiring a Senior Scientist-Host Microbe Interactions in Memphis, Tennessee. The lab studies the interactions between pathogenic bacteria and their hosts. Review all of the job details and apply today!
0
4
9
It’s my great pleasure to present the Global Pneumoocccal Sequencing project at #LISSSD25 I feel humbled to learn more about Group A and Group B streptococci and other species, and meet great minds to discuss science. #GPS_project
0
3
15
Dr Dorota Jamrozy presents a well-powered GWAS to identify genetic features associated with invasive #GBS disease as compared with carriage at #LISSSD25 Thanks to the >20,000 GBS genomes in the #JUNO_project @sangerinstitute
0
1
6
Over half a million of pneumococcal disease in children reported in China annually Our paper showed 1) 94% serotypes covered by PCV13 2) 99.8% isolates resistant to ≥1 antibiotics Routine PCV use in children will ↘️ pneumococcal disease and AMR https://t.co/bvVOvtshNW
microbiologyresearch.org
2
4
3
Good morning Brisbane! What a nice view to prepare my very first talk at #LISSSD25 ! Looking forward to meeting people at the conference. #GPS_project #pneumococci #Genomics
1
2
27
☑️ Attended my first Europneumo. ☑️ Presented a poster at a conference for the first time. ☑️ Had a great time 😁 Loved being able to share what I've been working on over the past 6 months about the global patterns of serotype 4.
0
2
9