John Lees
@johnlees6
Followers
1K
Following
181
Media
16
Statuses
280
Pathogen informatics and modelling https://t.co/2Qg0pvaFg3 (Research group leader at EMBL-EBI - https://t.co/lJdwj0idt0)
Joined December 2009
Have you ever thought of doing a sabbatical at EMBL? It’s a great opportunity for scientific exchange and growth, and for learning advanced technologies and top-notch science! Three scientists shared their experiences on their research visits at EMBL. 👇 https://t.co/2n6B6bfSQ0
0
13
20
If you're a a master's student studying at a French university interested in these we're a possible host group -- send me an email to discuss
Are you a student in France studying computer science, statistics, or bioinformatics? @FranceintheUK has opened a new round of internships hosted at EMBL-EBI. Find out more and apply. https://t.co/o9QdzfUKyn
0
6
13
A mere six years after its first release, SKA is now published https://t.co/vtJGt98ged
3
16
51
This is exactly the kind of thing we wanted to happen!
A cool upcoming feature: When you upload a sample to Solu Platform, we’ll search all public NCBI samples for related genomes. This is built on the awesome AllTheBacteria database by Martin Hunt, Leandro Lima, @shenwei356, @johnlees6 & @ZaminIqbal
2
6
41
This neat paper about rapid evolution of bacterial transformation rate by @eprocha @labXC & co takes some of its inspiration from an earlier @PLOSBiology paper by Nick Croucher & @ChristoPhraser:
journals.plos.org
Modelling and high-resolution genomic data demonstrate the evolutionary significance of an ongoing arms race between bacterial uptake of environmental DNA ("transformation") and horizontal DNA...
Why do #bacterial species' #TransformationRates vary so dramatically? @epcrocha @labXC &co show that #Legionella & #Acinetobacter transformation rates evolve by sudden changes, likely caused by conflicts between #MGEs and the host bacterium #PLOSBiology
https://t.co/odHj02Kjws
0
5
14
Daria Frolova's paper in plasmid relatedness now out in MGen! She uses rearrangement distances & shared seq content to build a relatedness network of plasmids, identifies hubs which are likely transposons, and then clusters related plasmids. https://t.co/fG0GCYqKZO
@MicrobioSoc
microbiologyresearch.org
First paper from my student Daria Frolova's PhD now on biorxiv: https://t.co/Ex2tbOujgG The key question: plasmids change structurally ~as fast as they mutate, so how can we decide whether 2 plasmids are "the same plasmid" for epi purposes? 1/n
4
27
67
Happy to announce that our (me and @YunWilliamYu) metagenome profiler sylph ( https://t.co/WGX3W1yOjT) is now published in @NatureBiotech! 1/5
github.com
ultrafast taxonomic profiling and genome querying for metagenomic samples by abundance-corrected minhash. - bluenote-1577/sylph
Rapid species-level metagenome profiling and containment estimation with sylph https://t.co/rgG2K0Li2N
7
48
130
Extension to ska 'skalo' -- using local assembly to recover indels and MNPs where split k-mers mismatch https://t.co/I4tilmrq3U We hope to integrate this into the main ska package in future too
biorxiv.org
Insertions and deletions (indels) are important contributors to the genetic diversity and evolution of pathogens like Mycobacterium tuberculosis . However, accurately identifying them from genomic...
0
6
24
I'm looking for a know fusion signature in a graph that contains annotated human transcripts (from Refseq) and samples, including prone to fusion (human carcinoma samples).
1
3
1
There is still time to register for our SMBE satellite meeting on pangenomes and transmission in Tokyo (until 31st October) See our website for more details, and now a provisional schedule too! https://t.co/HnKRbshoVD
We are happy to have been selected by SMBE to host a satellite meeting on Pathogen Pangenome Evolution Between and Within their Hosts: https://t.co/HnKRbshoVD We are expecting the meeting to be in Tokyo on 26-28th November 2024, save the date!
0
11
15
Also now old Celebrimbor is a big name in rings of power s2 maybe our backronym will finally land
0
0
6
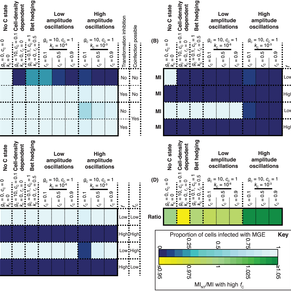
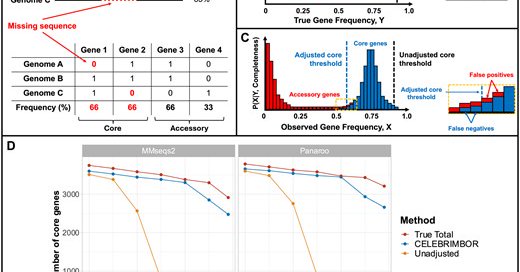
Now published in bioinformatics, CELEBRIMBOR 'pangenomes from metagenomes' https://t.co/L7FL5fekfe I think reviewer comments helped make it better, so worth another look (at fig 1 particularly!)
academic.oup.com
AbstractMotivation. Metagenome-Assembled Genomes (MAGs) or Single-cell Amplified Genomes (SAGs) are often incomplete, with sequences missing due to errors
Sam has then wrapped it all up in a nice snakemake pipeline CELEBRIMBOR (Core ELEment Bias Removal In Metagenome Binned ORthologs) which lets you go from MAGs to pangenome https://t.co/c3BZprY94E
1
18
50
Sam has then wrapped it all up in a nice snakemake pipeline CELEBRIMBOR (Core ELEment Bias Removal In Metagenome Binned ORthologs) which lets you go from MAGs to pangenome https://t.co/c3BZprY94E
1
2
2
📢 Only a week left to submit your abstract for #EMBLInfectionBiology 🦠 The event is a platform for late-stage postdoctoral scientists in infection biology to connect with the leading European institutions in the field. It is fully virtual for the ease of access 💻 Discuss
0
9
17
A reminder that abstract submission for the SMBE satellite meeting on pathogen pangenomes and transmission closes soon (8th September, a short extension). See: https://t.co/HnKRbshoVD
1
1
6
I have written up some new notes in the intermediate programming course on parallel programming, which are available here: https://t.co/SIdrMS8Rhn The main intro and advanced examples are in rust, but I've also added two independent sections covering python and R.
0
4
15
I was excited by a preprint on 'PanMAN' by @yatishturakhia and co, here's a mini-review on my initial thoughts and how it might apply to bacteria: https://t.co/q1vUKwAM57
johnlees.me
This is a mini-review (just highlighting some initial thoughts) of this preprint: Compressive Pangenomics Using Mutation-Annotated Networks Sumit Walia, Harsh Motwani, Kyle Smith, Russell Corbett-D...
1
3
12
🌞🌞🌞Job alert ‼️ We have not one but two positions available with me in @IneosOxford @OxfordBiology. More details here: https://t.co/nAW2DFaszm If you're interested, get in touch. Please share/RT #AMR
ineosoxford.ox.ac.uk
Join our cross-disciplinary team dedicated to finding solutions against AMR
5
47
62
Josie Bryant (@Josie_M_B) and I have a 3yr joint postdoc project at Sanger & EBI (ESPOD) Topic: Developing new bioinformatic and modelling approaches for exciting new longitudinal microbiome datasets Project details: https://t.co/MY9SaO8x33 To apply:
ebi.ac.uk
The ESPOD fellowship builds on the collaborative relationship between EMBL-EBI and the Wellcome Sanger Institute, offering projects that combine experimental (wet-lab) and computational (dry-lab)...
0
22
30
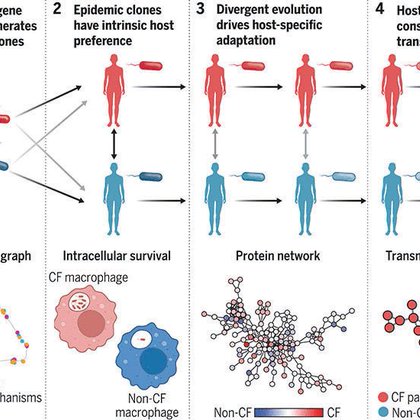
Today in @ScienceMagazine, we report how a bacteria has evolved into a major human pathogen over the last 200 years. Read on, this is going to be an adventurous ride 1/n!
science.org
The major human bacterial pathogen Pseudomonas aeruginosa causes multidrug-resistant infections in people with underlying immunodeficiencies or structural lung diseases such as cystic fibrosis (CF)....
11
171
598