Sachin Pundhir
@pundhir_sachin
Followers
191
Following
452
Media
72
Statuses
857
Senior Scientist - Computational biology @UCPH_BRIC @Rigshospitalet Interest: regulatory genomics in stem cell differentiation and cancer using #DataScience #ML
Copenhagen, Denmark
Joined May 2015
Invite you to read our latest work where we study the affect of SWI/SNF and NuRD inactivation on gene expression, which we find is clearly defined by RNA Pol II occupancy levels at gene promoters. https://t.co/Hvbsd6nlDQ with Jinyu Su @Bo_Porse @Marta12724018 🧵 1/6
1
5
19
👀 New tidyverse package being developed by @hadleywickham: elmer: Call LLM APIs from R https://t.co/waYyw92BfD
#Rstats
0
7
20
There is still a bit more than a week for people to apply 😉
🚨 PhD Opportunity Alert! 🚨 We are thrilled to offer a three-year, fully funded PhD position in Deep Generative Models for Single-Cell Sequencing Analysis, a joint project between Jes Frellsen's group and myself. https://t.co/9tB0vcvNkf
#PhDPosition #SingleCell #DeepLearning
0
3
3
Amazing thread showing, among other things, how DNA methylation can be a positive regulator of gene expression.. https://t.co/xJgzkoAzcn
Here is Max’s drawing about what we think is happening at the Zdbf2 locus, which really helped wrap my brain around these loops
1
0
2
🧵 Bioinformatics faces a critical challenge: despite its importance, software quality often falls short due to the lack of proper development practices. In our latest paper, we advocate for improving software quality in bioinformatics through teamwork. https://t.co/5PxmnDSiZx
3
47
184
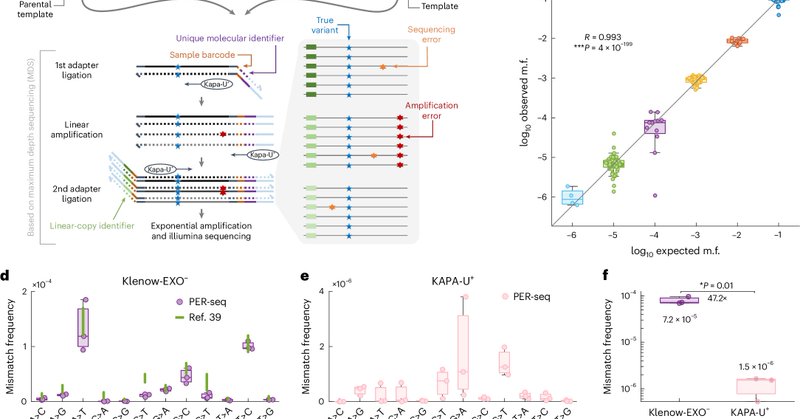
Cost of having DNA methylation: DNA replication by DNA polymerase epsilon is a source of C to T mutations in CpGs, especially methylated ones. This expands our understanding of one of the most widespread endogenous mutational mechanisms.
nature.com
Nature Genetics - A new method called polymerase error rate sequencing (PER-seq) can measure the nucleotide misincorporation rate of DNA polymerases. DNA polymerase ε mutants produce an excess...
7
76
216
Pharmacological restriction of genomic binding sites redirects PU.1 pioneer transcription factor activity https://t.co/Nj5o0YxwlA
nature.com
Nature Genetics - Chemically driven blockade of PU.1 binding sites leads to its genome-wide redistribution. PU.1 network rewiring causes human acute myeloid leukemia cells to differentiate.
0
1
4
Erasmus Virtual Hematology Lectures starts again on Monday September 2 at 16.00 CET! We have MAXIM PIMKIN from Harvard/Boston with: "Gene-regulatory networks in leukemia maintenance and immune escape". Link: https://t.co/ICOMFc5jv5 Spread the news! @pimkin
1
18
40
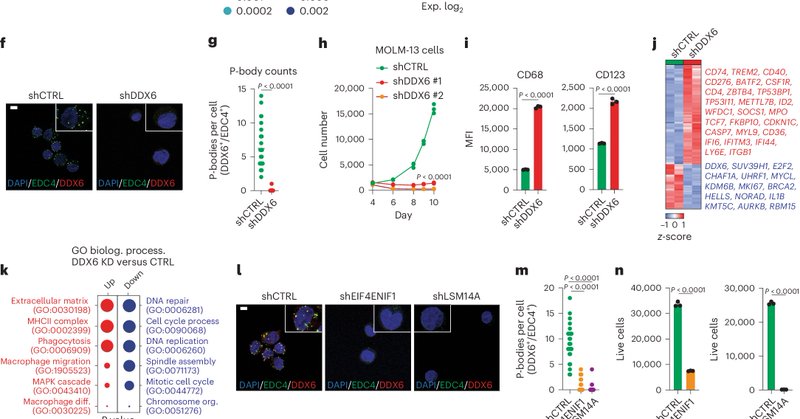
1/Excited to share our latest study showing that RNA sequestration in P-bodies sustains myeloid leukemia - now out in @NatureCellBio! https://t.co/3lIKztpOdg. 🧵 below:
nature.com
Nature Cell Biology - Kodali, Proietti et al. report that increased numbers of P-bodies in leukaemia cells account for sequestration and prevention of tumour-suppressive mRNAs from being...
14
26
150
Excited to share our latest work investigating the consequences of extant genetic variation in cis-regulatory elements at single-cell resolution. A short 🧵 https://t.co/fdq9U83Csf
biorxiv.org
Gene expression and complex phenotypes are determined by the activity of cis -regulatory elements. However, an understanding of how extant genetic variants affect cis -regulatory activity remains...
1
27
72
Woohoo - Our new paper is out! 🥳 https://t.co/0bPYvE2kts It offers new insight into the timing of DNA replication in embryos right after fertilization. My sincerest thanks to everyone involved 🙏 Here is a short thread on what we learned: 1/7
nature.com
Nature Communications - Here, the authors reveal the emerging replication timing program in mouse zygotes and 2-cell embryos, which differs from embryonic stem cells. They find systematic...
6
20
72
⚡️ Excited to share that I am starting an AI+Education company called Eureka Labs. The announcement: --- We are Eureka Labs and we are building a new kind of school that is AI native. How can we approach an ideal experience for learning something new? For example, in the case
2K
4K
28K
TFscope: systematic analysis of the sequence features involved in the binding preferences of transcription factors https://t.co/3mL5RU8iTJ
1
31
148
One week left to apply for our 2nd Statistical Genomics workshop, Sep 2-6, 2024 @NCBS_Bangalore! Registration details here: https://t.co/2VHi3I5YU7
Our 2nd statistical genomics workshop, Sep 2-6, 2024, jointly with @mkjolly15 @skouped, is open for registration. If you're an early-career researcher interested to participate, check the link below for more details https://t.co/2VHi3I5YU7 Registration deadline: 20th July 2024
0
32
75
Please see job advert for an exciting vacant PhD position in our group.
linkedin.com
We are looking for suitable candidates to fill an exciting vacant PhD position in our group at the Aarhus University, Department of Animal and Veterinary Sciences. Successful candidate will work in...
0
1
1
New paper in NatureComm: "Epigenetic alterations affecting hematopoietic regulatory networks as drivers of mixed myeloid/lymphoid leukemia". https://t.co/soC6QFRLpZ Great Rotterdam/Regensburg collaboration: @RogerMulet @ClaudiaGebhard Bas Wouters @DelwelRuud Michael Rehli
0
13
59
Alternative promoters drives key oncogenic processes during tumor progression, we showed it on liver cancer https://t.co/ZzssIwo3Yr Now similar observation in prostrate cancer https://t.co/M7OZoVSEOk Citation would have been appropriate. Pls look at alt-promoters in ur data.
nature.com
Nature Cell Biology - With ultra-deep RNA sequencing, Zhang et al. report increased usage of alternative promoters driven by AR, FOXA1 and MYC during prostate cancer progression and suggest altered...
0
4
13
How do enhancers work over distances that sometimes exceed megabases? Excited to share our work led by @gracecbower where we uncover a unique sequence signature globally associated with long-range enhancer-promoter interactions in developing limb buds: https://t.co/n0p7BdxSdq 1/
23
232
693