Peter Kulcsar
@p_kulcsar
Followers
24
Following
5
Media
0
Statuses
10
Post-doc at @UZH_en in @schwanklab, formerly PhD in @welkerlab // #CRISPR #geneediting
Joined October 2019
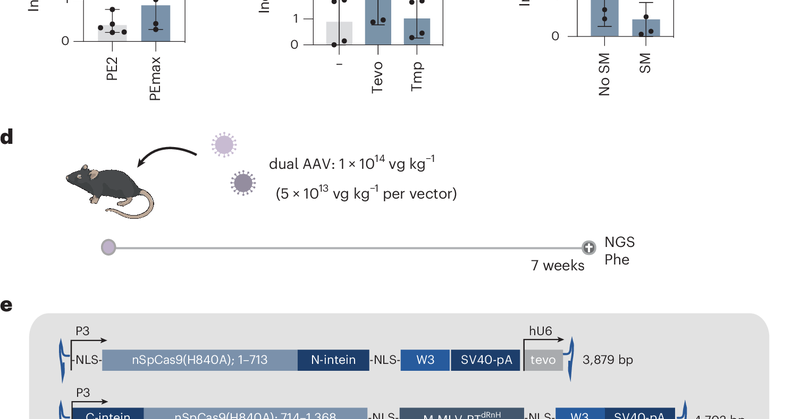
RT @talasandris: Our paper describing an RNA-LNP prime editing strategy for treating phenylketonuria was published today in @natBME (1/7).h….
nature.com
Nature Biomedical Engineering - A transient in vivo prime editing strategy is developed for the liver by delivering the prime editor as mRNA encapsulated in LNP.
0
10
0
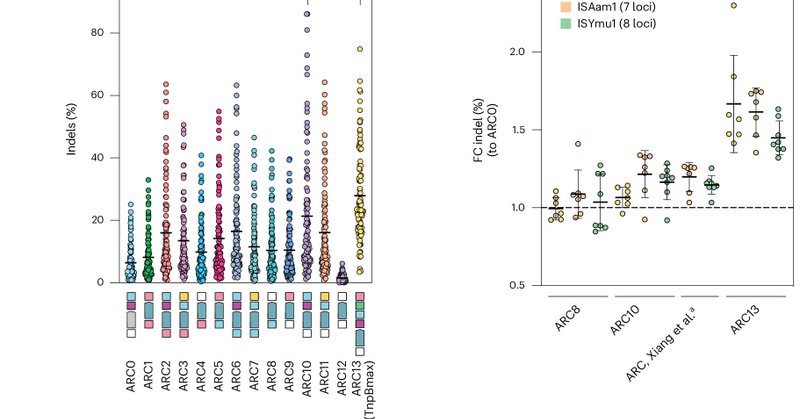
RT @schwanklab: Excited to share our latest study!.Today in @naturemethods the @schwanklab and @krauthammerlab report in @marquark et al. '….
nature.com
Nature Methods - This work introduces engineered TnpBmax proteins with enhanced efficiency and an expanded targeting range. By leveraging an extensive dataset on editing efficiencies, it also...
0
30
0
RT @schwanklab: Here we demonstrate that OrthoRep can be employed to evolve genome editors - Congrats to Yanik and many thanks to all colla….
0
22
0
Really nice and useful work done by my colleague @nicopmat! If you need to predict PE outcomes give it a try at @schwanklab.
🚨New preprint🚨Today, we're excited to share our latest study, "Predicting prime editing efficiency across diverse edit types and chromatin contexts with machine learning" via @biorxivpreprint! @schwanklab @krauthammerlab @bvansteensellab @UZH_Science.🧵.
0
0
6
RT @nicopmat: 🚨New preprint🚨Today, we're excited to share our latest study, "Predicting prime editing efficiency across diverse edit types….
biorxiv.org
Prime editing is a powerful genome editing technology, but its efficiency varies depending on the pegRNA design and target locus. Existing computational models for predicting prime editing rates are...
0
26
0
In our recent work published in @NatureComms we demonstrated a strategy to efficiently edit practically any target without off-targets using SpCas9 variants. The ‘CRISPRecise set' of variants will be available as a kit from @Addgene. @welkerlab @BiochemBrc
1
4
15
RT @welkerlab: In our new paper published in @NatureComms we characterize the on-target activity and off-target propensity of SuperFi-Cas9….
0
4
0
In our recent @biorxivpreprint, using target-matched SpCas9 variants, we demonstrated efficient editing without off-targets on any SpCas9 target sequence, even on challenging targets that were attempted but failed in all previous studies. @BiochemBrc
biorxiv.org
Streptococcus pyogenes Cas9 (SpCas9) has been employed as a genome engineering tool with a promising potential within therapeutics. However, its off- target effects pose a major limitation for...
0
3
6
RT @welkerlab: In our recent @biorxivpreprint we report that SuperFi-Cas9 exhibits extremely high fidelity but reduced activity in mammalia….
biorxiv.org
Several advances have been made to SpCas9, the most widely used CRISPR/Cas genome editing tool, to reduce its unwanted off-target effects. The most promising approach is the development of increased...
0
2
0
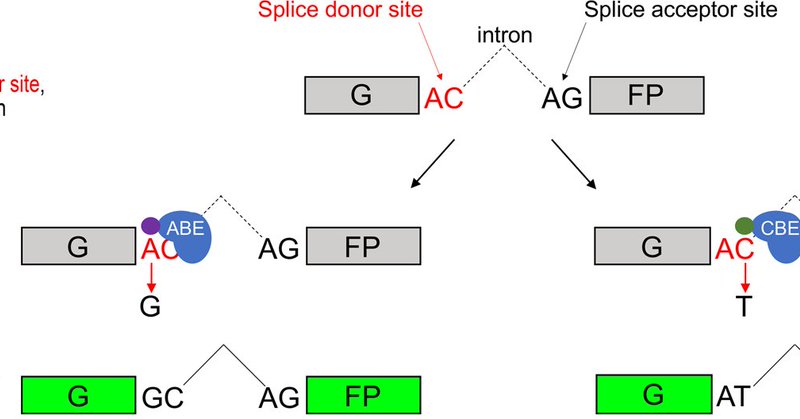
RT @welkerlab: We report in @NatureComms on the development of Base Editor Activity Reporter (BEAR) to study the mechanism of ABE and CBE v….
nature.com
Nature Communications - Base editors allow for precision engineering of the genome. Here, the authors present BEAR, a plasmid-based fluorescence assay for the measurement of CBE and ABE activity,...
0
3
0