OmniPath
@omnipathdb
Followers
399
Following
18
Media
27
Statuses
68
OmniPath: molecular biology knowledge from 100+ databases
Heidelberg, Germany
Joined October 2017
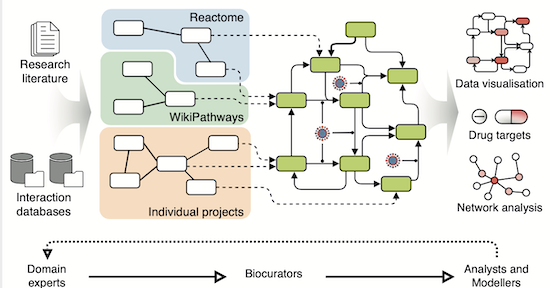
Building a knowledge graph for biomedical tasks usually takes months or years. 😰 What if you could do it in weeks or days? 🏃 We created BioCypher ( https://t.co/9eGe9RDkNj) to make it easier than ever, but still flexible and transparent. 🧵⬇️
3
107
319
Check out LIANA that uses @omnipathdb to systematically compare and combine resources & methods to study cell-cell communication from #scRNA
We are 🥳 to share that a substantially improved version of LIANA’s manuscript "Comparison of Resources and Methods to infer Cell-Cell Communication from Single-cell RNA Data" has now been published at: https://t.co/Tdy1MOkCPz Highlights for some of our findings in the 🧵1/10
0
0
3
We have also added several tutorials showing how to use decoupler and @omnipathdb in single cell analyses such as automatic cell type annotation, transcription factor and pathway activity inference. Documentation and tutorials available in: https://t.co/70IL1CZ4Ya
0
2
9
We are looking for PostDosc/ PhDs in Medical Bioinformatics & Machine Learning @uniklinik_hd @UniHeidelberg #multiomics #singlecell consider applying and pls RT 🙏
jobrxiv.org
Post a job in 3min, or find thousands of job offers like this one at jobRxiv!
1
56
73
Check MISTy from @tanevski et al @saezlab to analyze spatial omics - they used @omnipathdb 's annotated prior knowledge to link intracellular pathway activities & the expression of ligands in space 👇
Play MISTy for me, says someone with spatial omics data to find relationships between markers in. It's a machine learning method from @tanevski, @saezlab & co. It's demonstrated on mass cytometry and spatial transcriptomics data from breast cancer https://t.co/K9WbEg6vky
0
2
4
Our updated manuscript describing methods for bioactivity inference (pathways, TFs, etc.) is now published in @BioinfoAdv
https://t.co/x5beAKix7V 🥳 We have been working on a new #Python 🐍 implementation and its integration with the meta-resource @omnipathdb 👇
There are many computational methods to infer biological activities. Many lab members led by @PauBadiaM have developed decoupleR 💻, a #Bioconductor #rstats package that collects different methods to infer mechanistic signatures from omics 🧬 Paper: https://t.co/Swa6HqpfMo 1/10
3
26
49
The great spatial omics platform from @fabian_theis group is out in @naturemethods 🥳 Squidpy uses OmniPath data for ligand-receptor interactions 🤩 Special thanks to Michal Klein for implementing the OmniPath Python client✌️
Interested in spatial omics? Plz check out our squidpy tool, bringing together omics & image analysis to enable a scalable description of spatial molecular data. Out today @naturemethods thx to @g_palla1, @HannahSpitzer1 & team! https://t.co/0iD92imm4H
https://t.co/VPpelQHzl1
0
7
19
Our paper is among the 8 most read in @MolSystBiol
https://t.co/yBDMGshUmb, and got already 27 citations! 🎉 In @saezlab & @KorcsmarosLab it's a great pleasure for us to see you using our work. Meanwhile we are preparing big new developments, stay tuned 😉
0
10
29
Check out #COVID19 Disease Map Consortia @CovidPathways knowledge repository https://t.co/QHbzxHLoEJ, great community effort where we contributed @omnipathdb (pathway knowledge), DOROTHEA https://t.co/Z2V6XUvhV2 (TF targets) & CARNIVAL https://t.co/NJW3YjBKnh (network modeling)
link.springer.com
Molecular Systems Biology - We need to effectively combine the knowledge from surging literature with complex datasets to propose mechanistic models of SARS‐CoV‐2 infection, improving...
0
3
11
We just talked about OmnipathR at BioC2021 @Bioconductor, many thanks to all the participants and the organizers! 🌞 Slides: https://t.co/bAyvt0iNyN Vignette (and further materials): https://t.co/oSGIKD8Fx7 Other @saezlab tools:
0
1
3
11 #PPI scores based on structure, sequence and function from PrePPI of Honig Lab @Columbia 🌴 https://t.co/ksrM9XiTt1 🌴 Now easy access with our R package: https://t.co/knmgpILYDd 🌴 https://t.co/3faHuMPPGH
0
2
3
6,225 new protein complexes in OmniPath thanks to https://t.co/XwXFzXoZ1z 2.0 from @kdrew @jbwallingford & @edward_marcotte 🖥️ https://t.co/JzCOhnFH7B, 🏡 https://t.co/HQa61Q0PyZ, 📰 https://t.co/ZcOIfQsuzr
0
4
3
Human Cell Map from @gingraslab1 is available in OmniPath! ⭐️ Subcellular localization of 6,000+ proteins from a BioID screen in HEK cells: https://t.co/BRVn6U3Nge,
https://t.co/q9doPHOv0i
0
3
8
Estimate ligand activities from #transcriptomics with the new OmniPath-NicheNet workflow 🤩 https://t.co/7l5KZKWsp9 🤩 Customizable networks from OmniPath & other resources + NicheNet algorithm from @RobinBrowaeys of @saeyslab
0
9
34
Check out my first first-author work with @ManishButte! We used @omnipathdb, a protein interactions meta-database, as well as RNAseq data from @ProteinAtlas to expand the potential genes of inborn errors of immunity (IEIs). Hoping immunology clinics worldwide will find it useful!
Expanding the Potential Genes of Inborn Errors of Immunity through Protein Interactions https://t.co/fP0bkIWjOW
#bioRxiv
9
11
36
Happy that @omnipathdb was helpful as a meta-resource of cell-cell interaction resources in this comparison of methods to estimate cell-cell communication from scRNA-seq data
There are many tools and resources to estimate Cell-Cell Communication from scRNA data - do you wonder which one you should use? We do too! To find out, we compared 6 methods and 15 resources https://t.co/s7gwjK4Nne.
0
0
5
Access OmniPath from R https://t.co/3faHuMPPGH, Python https://t.co/4H55Osnpi6 and @cytoscape
https://t.co/GZubIM3ynf, or build your own database by pypath https://t.co/7RdJkM6gvc - and send us feedback and suggestions
0
3
3
OmniPath provides prior knowledge for many omics analysis and modeling tools, including a close integration with NicheNet @saeyslab to infer ligand activities from transcriptomics and the single-cell platforms squidpy + @scanpy_team @fabian_theis
https://t.co/mqIKHSCLww
1
1
5
Our case studies about autocrine signaling in #SARS_CoV_2 infected Calu3 cells @alvaldeolivas and myofibroblast-Treg cell signaling in #IBD (ulcerative colitis) with @KorcsmarosLab show how OmniPath helps to analyze intercellular communication
1
3
2
Protein function, disease role, tissue expression, localization, structure and more in the annotations database from 50 resources
1
1
2