Loyal Goff
@loyalgoff
Followers
1K
Following
3K
Media
43
Statuses
2K
Associate Professor @ JHU, explorer of single (brain) cells, chaotic/good, Dad^^2, My contributions to science not defined by my position on author lists.
Baltimore, MD
Joined November 2007
Pleased to present 'tricycle', a new R package for rapid, accurate, and scalable estimation of cell cycle position for scRNA-Seq using transfer learning. A joint project from our lab and the lab of @KasperDHansen.
1
35
102
RT @ShechnerLab: It's my pleasure to present the next big preprint from SheqLab! An exciting application of our O-MAP platform that I hope….
0
51
0
RT @genandgenes: Heading to #ASHG24 and looking for a postdoc position? . Interested in genetic epidemiology, specifically gene x environme….
0
19
0
RT @DongeunHeo: My second manuscript from graduate school is now on bioRxiv! We generated a comprehensive scRNA-seq dataset of mouse OPCs f….
0
17
0
RT @TheSurajKannan: Without getting into the science itself, I wish everyone would read the editor's statement for the review of this manus….
elifesciences.org
Mesoderm-derived enteric neurons born in the post-natal vertebrates drive maturation and aging of the enteric nervous system.
0
1
0
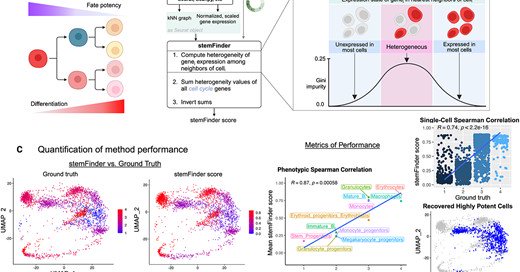
RT @ekernf01: I want to boost the freshly published main PhD work of a @cahanLab colleague, Kathleen Noller, whose thoroughness and work e….
academic.oup.com
Abstract. Methods that predict fate potential or degree of differentiation from transcriptomic data have identified rare progenitor populations and uncover
0
6
0
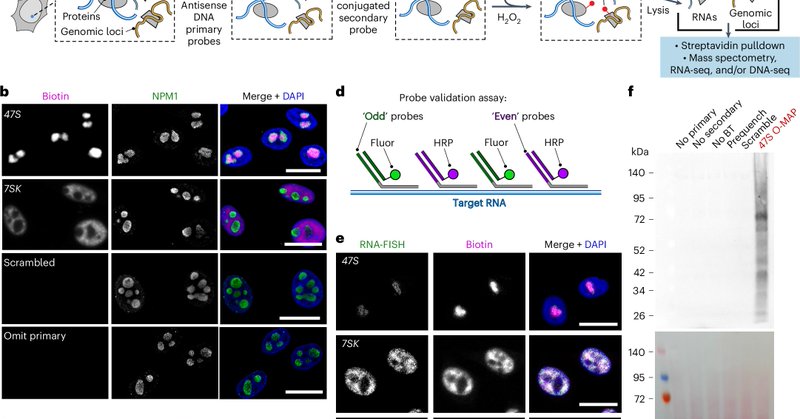
RT @ShechnerLab: Beyond thrilled to present our O-MAP—now *published* @naturemethods (!!!). O-MAP is a powerful new method for biochemicall….
nature.com
Nature Methods - Oligonucleotide-mediated proximity-interactome MAPping (O-MAP) enables precise multiomic characterization of biomolecular interaction networks at target RNAs. Distinct O-MAP...
0
110
0
RT @ctmurphy1: The LSI at Princeton University invites applications for a tenure-track faculty position at the Assistant Professor level. W….
0
119
0
RT @WiringTheBrain: A lot of molecular and cell and developmental biology is concerned with figuring out the "functions" of various protein….
0
140
0
RT @lpachter: This recently published figure by @Sarah_E_Ancheta et al. is very disturbing and should lead to some deep introspection in th….
0
94
0
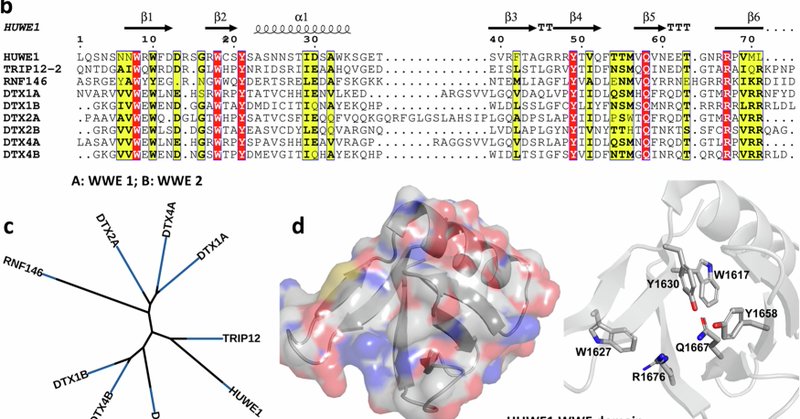
RT @LeungLab: Happy to be part of a multi-institutional effort through an academia-industry partnership in the systematic analysis of the W….
nature.com
Communications Biology - Six WWE domain-containing proteins also contain domains with E3 ubiquitin ligase activity. We report structures, biophysical and biochemical assays for the discovery of...
0
13
0
RT @TheSilverLab: I am excited to share our most recent story led by our fantastic postdoc Lucas Serdar. How is steady state gene expressio….
biorxiv.org
RNA expression levels are controlled by the complementary processes of synthesis and degradation. Although mis-regulation of RNA turnover is linked to neurodevelopmental disorders, how it contributes...
0
16
0
RT @xinjin: 📢 Our latest paper is out in @CellCellPress! Led by the multi-talented @XinheZheng, we accelerated AAV-based gene delivery to 2….
0
195
0
RT @MBLScience: We're excited to announce this year's Whitman Fellows! The MBL will welcome 29 scientists from 27 universities and research….
mbl.edu
0
7
0
RT @MBLScience: We're excited to announce this year's Whitman Fellows! The MBL will welcome 29 scientists from 27 universities and research….
0
6
0
RT @pskatz: We are halfway through the first ever Cephalopod Neuroscience Conference at the MBL. Over 200 participants. What an amazing exp….
0
17
0
RT @sinabooeshaghi: Our paper "A machine readable specification for genomics assays" is now published in Bioinformatics, @OUPBioinfo. In sh….
0
38
0
RT @satijalab: As my lab develops Seurat, I wanted to share some thoughts on the important Seurat and scanpy package selection preprint fr….
0
212
0
RT @lpachter: It's been great to see the positive response of @satijalab & @fabian_theis to our preprint on Seurat….
biorxiv.org
Standard single-cell RNA-sequencing analysis (scRNA-seq) workflows consist of converting raw read data into cell-gene count matrices through sequence alignment, followed by analyses including...
0
58
0
RT @IgorUlitsky: 1. (by a mile) Reviewers asking for mechanism in papers describing functions/phenotypes. LncRNA mechanisms are very comple….
0
5
0
RT @IgorUlitsky: Since the seasonal dunking-on-lncRNAs is back, and I do 💕 lncRNAs, a 🧵 on what I think have been the sources of the main p….
0
32
0