Sarah Ancheta
@Sarah_E_Ancheta
Followers
104
Following
11
Media
6
Statuses
11
Associate Computational Biologist @czbiohub SF, incoming PhD student @UCSF #CZBiohubSF #RNAvelocity #zebrahub
San Francisco, CA
Joined March 2023
RT @loicaroyer: 🚨Big news! Five years in the making, our Zebrahub paper is now published in #Cell 🎉. We’ve built a timecourse atlas of zebr….
0
155
0
RT @biorxiv_bioinfo: Challenges and Progress in RNA Velocity: Comparative Analysis Across Multiple Biological Contexts .
biorxiv.org
Single-cell RNA sequencing is revolutionizing our understanding of cell state dynamics, allowing researchers to observe the progression of individual cells’ transcriptomic profiles over time. Among...
0
11
0
8/8 Read the full preprint to dive deeper into our methods and results. Your feedback and questions are welcome! Many thanks for the amazing mentorship from @Merlin_Lange, @ale_agranados, @sculptorofdance and @loicaroyer #AcademicTwitter #RNAvelocity #ScienceTwitter.
0
0
1
7/8 The choice of RNA velocity method can significantly impact the insights gained from scRNA-seq data. We hope our benchmark helps researchers choose the best tool for their data. Many thanks to #CZBiohubSF for their support! @czbiohub #OpenScience #Bioinformatics.
1
1
2
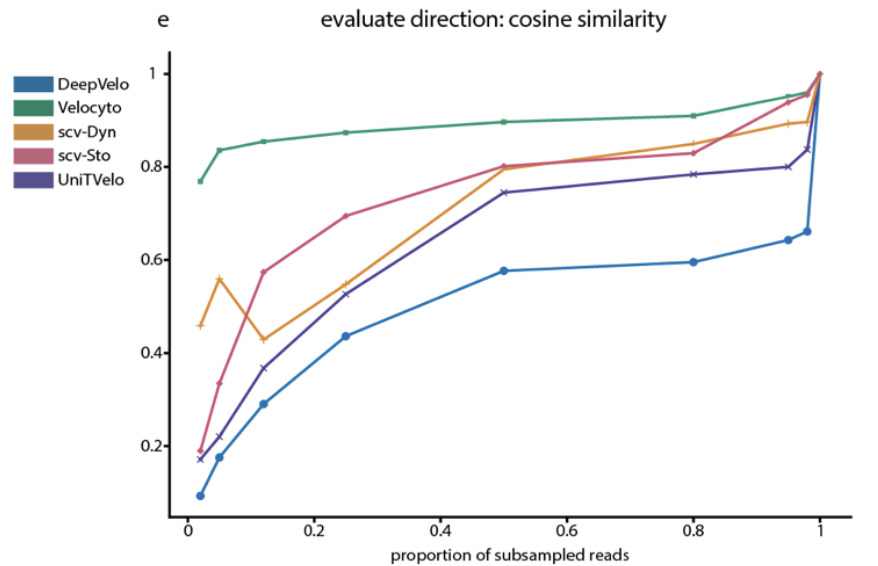
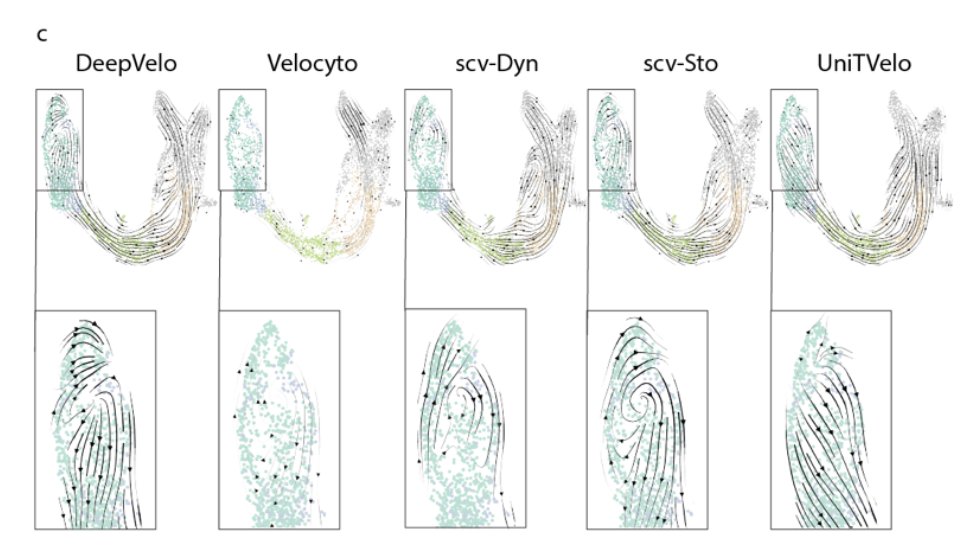
4/8 We observed significant discrepancies in RNA velocity predictions among different methods. We explore the agreement (or lack thereof) between methods for the zebrafish neuromesodermal progenitor dataset. #Bioinformatics #zebrahub
1
0
1
2/8 We analyze the performance of five RNA velocity methods across three developmental contexts. RNA velocity methods vary significantly in their predictions. Understanding these differences is crucial for trajectory inference. #single-cell #devbio #zebrafish #pancreas
1
0
1
1/8 Excited to share our preprint on RNA velocity! 🚀🧬 In this study, we compare 5 RNA velocity methods across 3 datasets. Check out our findings! #scRNAseq #RNAvelocity 📄
2
16
71
RT @loicaroyer: We present #Zebrahub: a timecourse atlas of zebrafish embryonic development, combining #scRNAseq time-course data with #lig….
0
170
0