Christopher Hoel
@chrismhoel
Followers
668
Following
13K
Media
30
Statuses
470
Inspired by @owl_posting's recent Socratic dialogues about the utility of DNA models, I wrote a Gödel, Escher, Bach-style dialogue about different approaches to protein–ligand binding-affinity prediction. (The weirdest writing I've shared publicly?) https://t.co/n7nHOPiaf1
corinwagen.github.io
A Hofstadter-style dialogue about protein–ligand binding-affinity prediction.
12
20
123
With the start of my first batch as a visiting partner at @ycombinator, this felt like a good time to write my founder story about our journey at @reverielabs. It's a bit of a long one. https://t.co/KWyMxPDdXO
ankitg.me
Lessons on Building an AI x Bio Startup
22
28
245
In new work led by @aditimerch with @samuelhking, we prompt engineer Evo to perform function-guided protein design with high experimental success rates, including designs that go beyond natural sequences. We also release SynGenome, the first AI-generated genomics database. 🧵 1/N
9
126
503
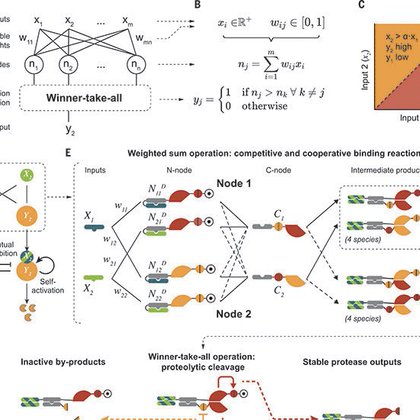
Excited to finally share Perceptein, a PERCEPtron made of proTEINs:
science.org
Artificial neural networks provide a powerful paradigm for nonbiological information processing. To understand whether similar principles could enable computation within living cells, we combined de...
9
50
199
PROTEIN DESIGN COMPETITION ROUND 2 The long awaited results are here -- and they don’t disappoint! 📈 We doubled the number of proteins we tested in our lab from 200 to 400! 🧬 Out of those 400 proteins, 378 expressed (95% expression rate!) 🚀 Out of those 378 expressed
4
55
201
Introducing ESM Cambrian. Unsupervised learning can invert biology at scale to reveal the hidden structure of the natural world. We’ve scaled up compute and data to train a new generation of protein language models. ESM C defines a new state of the art for protein
26
166
650
Can a single small molecule rescue the stability of nearly all mutations in a protein? Our new preprint by @taylor_mighell + thread https://t.co/pYw0RHCpKB
biorxiv.org
Reduced protein stability is the most frequent mechanism by which rare missense variants cause disease. A promising therapeutic avenue for treating destabilizing variants is pharmacological chapero...
A major challenge in treating rare genetic diseases is the huge number of causal variants in different individuals. This led to assumptions that any given treatment would be suitable only for a small fraction of patients
1
14
70
Remember Golden Gate Claude? @etowah0 and I have been working on applying the same mechanistic interpretability techniques to protein language models. We found lots of features and they’re... pretty weird? 🧵
21
191
1K
Have you ever wanted to design protein binders with ease? Today we present 𝑩𝒊𝒏𝒅𝑪𝒓𝒂𝒇𝒕, a user-friendly and open-source pipeline that allows to anyone to create protein binders de novo with high experimental success rates. @befcorreia @sokrypton
https://t.co/IPhMFpRgHh
26
436
2K
The Adaptyv results are out! Very few designs actually bound. The Design Process text is interesting, e.g., looks like the highest scoring was due to a "new user-friendly binder design pipeline based on AlphaFold2 backpropagation" from @MartinPacesa
🧬 Protein Design Competition Update 🧬 Experimental characterization for all protein designs has been completed! We are still processing the last experiment runs and will be releasing the results on Friday Sep 20 at 6 PM CET - 2 PM EST - 9 AM PDT https://t.co/QvTEyKd7ak
0
11
37
We've started IND-enabling studies on a small molecule corrector for Rho-associated autosomal dominant retinitis pigmentosa (Rho-adRP). I thought it would be fun to spend some time walking through our approach and why we are excited. Long 🧵1/
25
71
419
EvolutionaryScale is partnering with @Lux_Capital & @envedabio to bring you a Bio x ML Hackathon from Oct 10-20. Build on ESM3 *98 billion* and proprietary datasets of protein activity to develop next-gen bio applications. Open globally. Get started:
1
26
97
1/ I did my own little hackathon last weekend designing EGFR binders for @adaptyvbio's protein design competition. I was really excited to see that my submissions took the top 10 spots in the virtual scoring phase! I got some DMs asking about my process so here's a thread:
6
62
345
De novo protein design is great, but nature has millions of proteins- why not repurpose them? Introducing Raygun, a new approach to protein design. It allows you to miniaturize, magnify or modify any protein. We synthesized miniaturized variants of eGFP and mCherry! 1/
26
216
972
I've noticed that biotech cos built around a computational discovery platform have a different philosophy on company building and how to use software to gain sustainable advantage vs. 'traditional' biotech or pharma. I reflect on this in a new blog post:
atelfo.github.io
Reflections on the differing philosophies of company building between biotechs with and without a core computational discovery platform
7
36
233
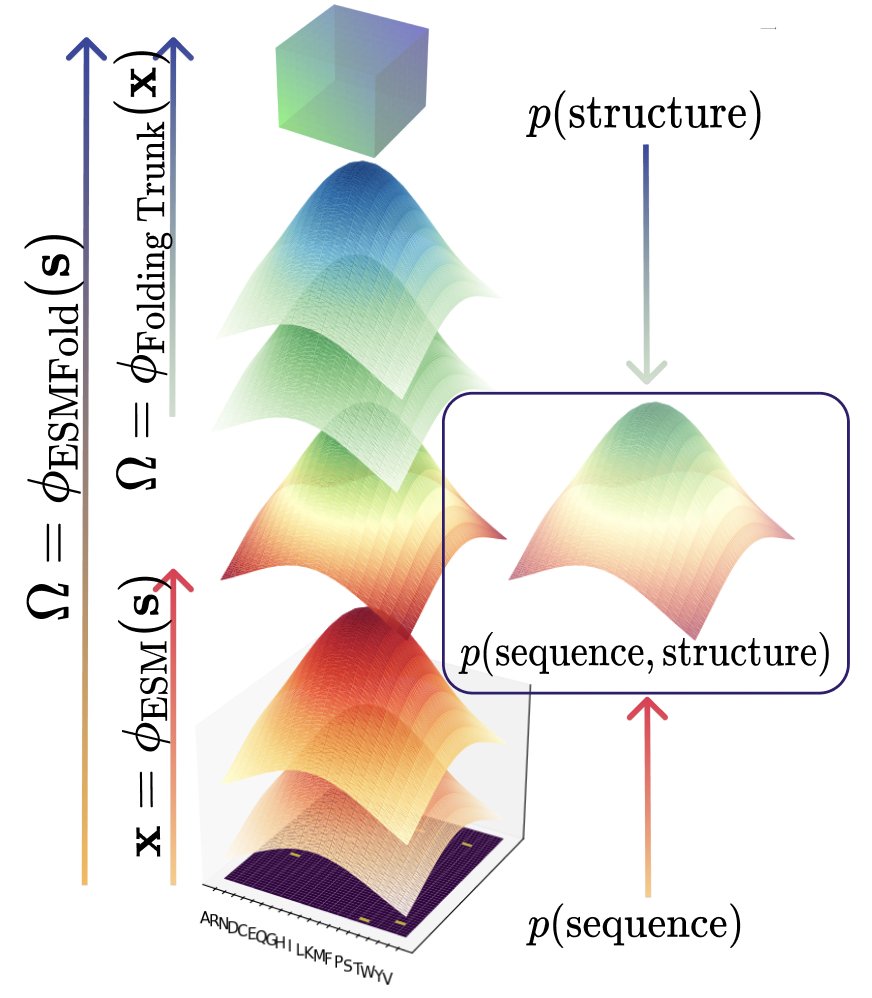
1/ 🧬 Excited to share CHEAP, our new work on compressed protein embeddings. We characterize the joint distribution of p(sequence, structure) in ESMFold's latent space, and find cool tidbits on compressibility, tokenizability, and pathologies: https://t.co/SkLCd2jBXn 🧵
6
98
473
The “known protein universe” is big: hundreds of millions of sequences from thousands of species. @ArcadiaScience we wondered, how well does it reflect the “actual” protein universe? 🧵[1/15] https://t.co/p4el3tmkx2
research.arcadiascience.com
Many protein prediction and design models rely on evolutionary comparisons. We show that popular databases are phylogenetically biased, influencing the statistical utility of the known protein...
2
30
105
We’re thrilled to share our new pre-print exploring how we can design and control dynamic proteins! (1/6) https://t.co/w7sRsdJV9y
5
116
509
One of the unforgettable moments at the 2024 Ion Channels GRC was the closing lecture by Fred Sigworth. If you missed it, here it is..
1
44
132