Chandler Sutherland
@chandler_suth
Followers
298
Following
784
Media
13
Statuses
115
Plant Biology PhD candidate @PMB_berkeley 🌱
Berkeley, CA
Joined May 2019
Diversity of brassica oleracea (plus a sneaky brassica rappa) for undergrad discussion section on crop breeding and domestication. I’ll also be eating very healthy this week
10
45
352
🧵 🥬 Lettuce begin! Super excited to share our new manuscript on contrasting patterns of evolution in sensor and helper NLRs in the NRC network! Tune in for some plant immunology and molecular evolution… 🌱🧬💻 (1/14) https://t.co/54sR517kEP
biorxiv.org
Nucleotide-binding domain and leucine-rich repeat immune receptors (NLRs) are known for their rapid evolution, even at the intraspecific level, yet the rates of evolution differ significantly across...
5
47
120
Natural variation modifies centromere proximal meiotic crossover frequency and segregation distortion in Arabidopsis thaliana https://t.co/27yUQwIuw8 new work from us on centromere inheritance and organisation @njgorringe @MpNaish @malacopa_genome
4
36
81
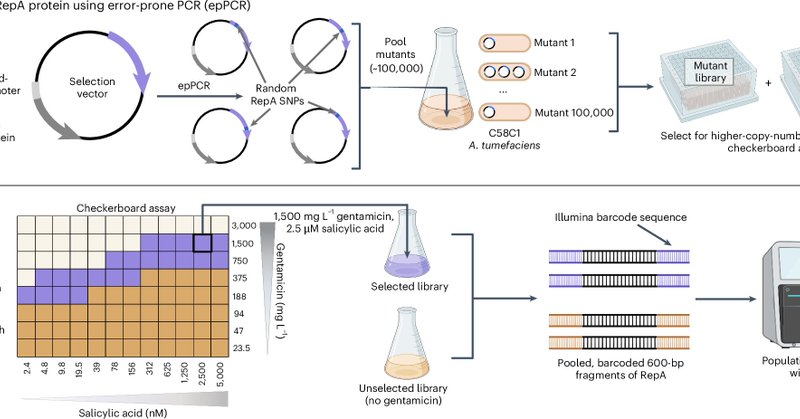
I am pleased to announce new research from the Shih Lab (@LabShih) published today in Nature Biotechnology on improving Agrobacterium-mediated transformation via binary vector copy number engineering ( https://t.co/nCUVQ2ym2p). 🧵A summary thread below🌿
nature.com
Nature Biotechnology - Agrobacterium-mediated transformation of plants and fungi is enhanced by plasmid copy number variants.
5
76
263
🦍Postdoc in Somatic Evolutionary Genomics🦍 Join my team @GeneticsCam on a pioneering project mapping somatic mutations across primates! 🌍Collaborate with experts @UCBerkeley & @sangerinstitute. 🔍Experience in genomics? Apply by Nov 16! Details: https://t.co/1pDO1tzzcP
0
74
125
I had so much fun on this UK Extravaganza!! Thank you to all our hosts and to everyone who shared their exciting science, see you soon🤩
It’s been great to be back to the UK, to catch up with colleagues and to meet new people. Thank you all for hosting and @chandler_suth for being a fantastic travel companion. Chandler giving a seminar - sketch by @ATJCagan
0
4
39
We had the pleasure to host @kseniakrasileva and @chandler_suth at @uclbiosciences at the beggining of their tour!
The finale of the Ksenia & Chandler “The NLRs Tour” is a joint @EarlhamInst @TheSainsburyLab Seminar cc @kseniakrasileva @chandler_suth
0
4
14
Published version @NatureEcoEvo of our collaborative work led by Patricia Lang @plantricia with @MExpositoAlonso and @stanfordstomata Labs https://t.co/7gIXzH3qdy
@MPI_Bio @CloeFor @uclbiosciences @UCLLifeSciences #herbariumgenomics to understand plant responses to #climatechange
1
10
38
A new preprint from @kseniakrasileva lab: Engineering pathogen-inducible promoters for conferring disease resistance in tomato
biorxiv.org
Plant diseases pose a significant threat to global crop production. Most disease resistance genes used in crop breeding programs encode nucleotide-binding leucine-rich repeat receptors (NLRs) that...
3
31
47
PMB is hiring a tenure-track Assistant Professor in Plant Developmental Biology! Apply by Oct 15, with expected start date of July 2025. For more information about the position, including required qualifications and application materials, visit https://t.co/g3W6J1mT9h.
0
31
31
We are excited to share our updated manuscript for our NLRseek program! This is a unique partnership between @2Blades, @TheSainsburyLab, and the Kaneka Corporation. Thank you to our co-authors for this success @MatthewMoscou
@PeterVanesse
@kamiljwitek
@UMNPlantPath Results: 🧵
Discovery of functional NLRs using expression level, high-throughput transformation, and large-scale phenotyping https://t.co/EsSYtmMz8C
#biorxiv_plants
3
36
78
🧵Excited to share my first author preprint detailing much of my dissertation work! Here we used gene editing to retrace the horizontal transfer of a bacterial gene that now encodes an anti-parasitoid toxin in Drosophila: https://t.co/KRKvzCIewb /1
biorxiv.org
Immune systems are among the most dynamically evolving traits across the tree of life, and long-lived macroparasites play an outsized role in shaping animal immunity. Even without adaptive immunity,...
10
22
77
Excited to share our recent work on annelid comparative genomics, led by @Tom_Lewin. Using @darwintreelife data, we assembled a dataset of 23 genomes and discovered lineage-specific chromosome scrambling. 1/3 https://t.co/WHxq4a3ZCZ
4
63
198
We have opened sign ups for our annual "Genomes and Plant Health" workshop that includes lectures and hands on activities. It is free, in person, Berkeley campus, June 10-11 undergraduates, June 12-13 high School students. Help us spread the word:
0
26
23
Why do some genome regions mutate more than others? Excited to share our latest work published in @ThePlantCell. H3K4me1 recruitment of DNA repair proteins in plants! 🌱🧬#PlantScience #Mutation 📜 https://t.co/bYqNTe6iqO 1/
2
41
149
Please RT - we are seeking a new postdoc! The Williams lab at UC Berkeley ( https://t.co/JmC7IPexrb) has a post-doc position available to study the impact of epigenetics on organ regeneration in plants.
williamsplantlab.com
Plant genetics & epigenetics at the Williams lab, Department of Plant & Microbial Biology, UC Berkeley
2
82
105
#BoG24 Diana Aguilar Gomez is telling us about genetic rescue in real time in the Florida Pumas - a very cool study of inbreeding, selection, and admixture @lohmueller
2
1
37
Majority of the highly variable NLRs in maize share genomic location and contain additional target-binding domains
biorxiv.org
Nucleotide-binding, Leucine Rich Repeat proteins (NLRs) are a major class of immune receptors in plants. NLRs include both conserved and rapidly evolving members, however their evolutionary traject...
0
8
28
Big thank you to everyone who provided feedback and guidance on this work, and to my co-authors @daniilprigozhin @kseniakrasileva @Grey_Monroe. Excited to start measuring mutation directly and hopefully get to some mechanistic answers soon🧬
0
1
7
Highly variable NLRs have distinct genomic features, selective pressures, and mutational likelihood contributing to their super high amino acid diversity. But what's the mechanism??? How are these features established, and how do they change local mutation probability??
1
2
4